Team:University of Washington/Project
From 2008.igem.org
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook | Protocols | BioBrick Parts Used | Glycerol Stocks |
|---|
Contents |
Controlled Trans-Kingdom Genetic Transfer
Bacteria exhibit a means of transferring genetic material separate from traditional generational inheritance. This phenomenon of horizontal gene transfer via bacterial machinery—termed conjugation—contrasts in many aspects with traditional heritable genetics. Specifically, the mechanism of bacterial conjugation does not rely on the transfer of genetic information from parent to progeny. Instead, a unique type of plasmid—called a mobile plasmid—induces the replication and transmission of itself and other plasmids between bacteria via direct cell contact. The genes on the mobile plasmid encode for a suite of conjugative bacterial proteins, including a sex pilus nanotube that connects the bacteria and accommodates the transmission of DNA. In this manner, bacteria can accept and express genes separate from their chromosomal DNA. While this mode of genetic transfer is fascinating on many levels, including its possible role in evolution and phylogenic discrepancies, it has the potential to be utilized as an invaluable mechanism for genetic alteration.
It has been demonstrated that bacteria can transfer genetic material across kingdoms to the eukaryote yeast, an organism incredibly evolutionarily distant from any bacterial species [1]. Unlike sexual reproduction, which requires great genetic similarity between mating partners, it would appear that conjugation provides a means for transferring genetic material across vast evolutionary expanses. Our goal is to engineer E. coli to perform trans-kingdom conjugation under the presence and absence of specific environmental signals. However, in order to prevent the genetic circuitry from being “evolved out,” the yeast plasmid will confer some adaptive function to the yeast. Eventually, the yeast will be engineered to produce some beneficial product for E. coli, and the two species will live symbiotically, exchanging DNA and cellular products necessary for survival. In this way, there will be a mutualistic interaction between the yeast and E. coli, and it is hoped that the functional integrity of the genetic circuitry will remain intact.
The modular design for trans-kingdom genetic transfer could be adapted for use with various organisms under different circumstances. However, our particular design is intended for a scenario where E. coli and yeast are in close proximity, the yeast cannot naturally digest lactose, lactose is the primary food source, and the E. coli possess a transferrable plasmid containing genes for lactose digestion. Because expending resources on lactose machinery in the presence of the more readily usable glucose would be disadvantageous, E. coli will only produce the LuxR protein in the presence of lactose, and the absence of glucose. The yeast will be engineered to produce the quorum sensing molecule, AHL. When the AHL diffuses to the LuxR proteins contained in E. coli, the production of conjugation machinery will be activated, and genetic material will be transferred to yeast. Pragmatically, this schema for trans-kingdom genetic transfer could be used to modify and engineer organisms which are normally difficult to access or operate on, and also as a means of delivering functional DNA for gene therapy. Additionally, this design may serve as an entry point for more complex, multi-organism systems.
LuxR Controls
Regulators: pLac
Lactose is a disaccharide composed of glucose and galactose, and in order for an organism to metabolize lactose, it first must be broken-up into these constituent monomers. β-galactosidase is an enzyme capable of doing just that, and is naturally produced by E. coli. However, E. coli cleverly regulate their production of this enzyme to times only when it’s required. Being that lactose is a less efficient fuel than simple glucose, E. coli only want to metabolize lactose when glucose is absent. This regulation is carried out at the transcriptional level by putting the β-galactosidase gene (and 2 other genes) under the control of a promoter (the lac promoter) which is inhibited by LacI and induced by CAP. LacI is a molecule that is constitutively produced by E. coli; it can either bind to the promoter or bind to lactose, meaning that LacI only inhibits the promoter in the absence of lactose. CAP is another molecule naturally present in E. coli which is only free to bind to the lac promoter in the absence of glucose. Given that gene repression is stronger than activation, the production of β-galactosidase is only carried out when required.
In order to have LuxR produced only when glucose is absent and lactose is present, the LuxR protein generator biobrick (part BBa_I0462) was placed under the control of the lac promoter (part BBa_R0010). The lac promoter is a commonly used transcriptional regulator, as it enables genes to be turned on/off with addition/subtraction of lactose (or IPTG) from the growth medium. On the other hand, the use of glucose as a means to regulate the activity of the promoter is not common. Thus, promoter part BBa_R0010 had to be tested to verify that it indeed behaves as the natural lac promoter does (i.e. IPTG can only successfully activate the promoter in the absence of glucose).
To carry out such a test, part BBa_R0010 was placed in front of the GFP generator biobrick (part BBa_E0240). An assay was then carried out that characterized the promoter activity in varying concentrations of IPTG and glucose: 25 wells of a 96-well plate were filled with the mid-log culture of a LacI expressing E. coli strain (DH5a+LacIq) grown in M9 minimal media. IPTG and glucose was then added to the wells in different amounts. The GFP fluorescence of each well was then measured over a six hour period. As the plot shows below, GFP has the highest expression in the well with the highest concentration of IPTG and no glucose, and the lowest expression in the well with the highest glucose concentration and no IPTG. Additionally, in wells with both glucose and IPTG, the repression of the glucose proved stronger than the activation of the IPTG, thus behaving just as needed.
Regulators: AraC and TetR
Since Lactose and Glucose also involve in cellular metabolism, there may be some complication that could affect our genetic circuits or how cells function in general. To try avoiding problems, we also make alternative promoter with AraC and TetR regulators to control LuxR production in E.coli. AraC is an activator induced by Arabinose sugar. TetR is a repressor that is made inactive by tetracycline(tc) or anhydrotetracycline(aTc).
In the research [http://www.nature.com/msb/journal/v3/n1/full/msb4100187.html "Programming gene expression with combinatorial promoters"] by Robert Sidney Cox, III, Michael G Surette, and Michael B Elowitz, it was shown that having AraC at distal region(upstream -35) and TetR at proximal region(downstream -10) made the most AND-gate promoter among the combination of AraC and TetR hybrid regulators.
The first approach to make the promoter is to insert TetR binding sites into existing AraC promoter biobrick (BBa_R0080) by QuikChange Mutagenesis method. This approach did not work out. Some possible reasons were that the primers were not well designed (i.e. homology of primers and parental plasmid were not long enough for primers to bind to parental plasmid.), and the materials were not correct or effective(reactions were made from existing lab's material without a kit).
The second approach is to PCR amplify the promoter region containing AraC and TetR from the clone D29 received from Elowitz's lab. Using primers with biobrick prefix and suffix hangings, we succeed biobricking the promoter part. The promoter is then put in front of LuxR.
Bacterial Conjugation
To test the AHL regulation of conjugation we are using lambda red recombineering to mutate the conjugation plasmid. This is being worked on in two separate ways on three different genes.
The major genes in regulating IncP conjugation are KorA, a global regulation transcriptional factor on the RP4 plasmid; TrbA, a transcriptional factor for the TrbB operon, which contains the essential genes for building the conjugation pili and other parts of the conjugation tube; and TrbC, the gene that produces the pilin protein.
Using the lambda red recombination plasmid, pKD78, we are going to create three knock out plasmids: KorA-, TrbC-, and TrbA-. These genes will then be placed on a secondary plasmid under the LuxR promoter, and we will test for conjugation in the presence and absence of AHL. Additionally, we plan to create two mutant IncP alpha plasmids with mutations in the wild type promoters of each transcriptional factor. Using lambda red, we plan to replace the wild type KorA promoter with the LuxR promoter and likewise with TrbA.
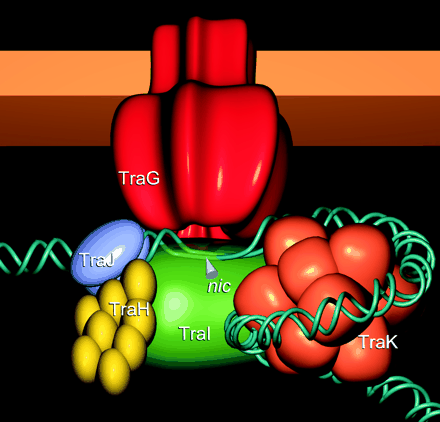
Illustration of Tra1 Conjugation Machinery
Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [http://www.ncbi.nlm.nih.gov/pubmed/11976307].
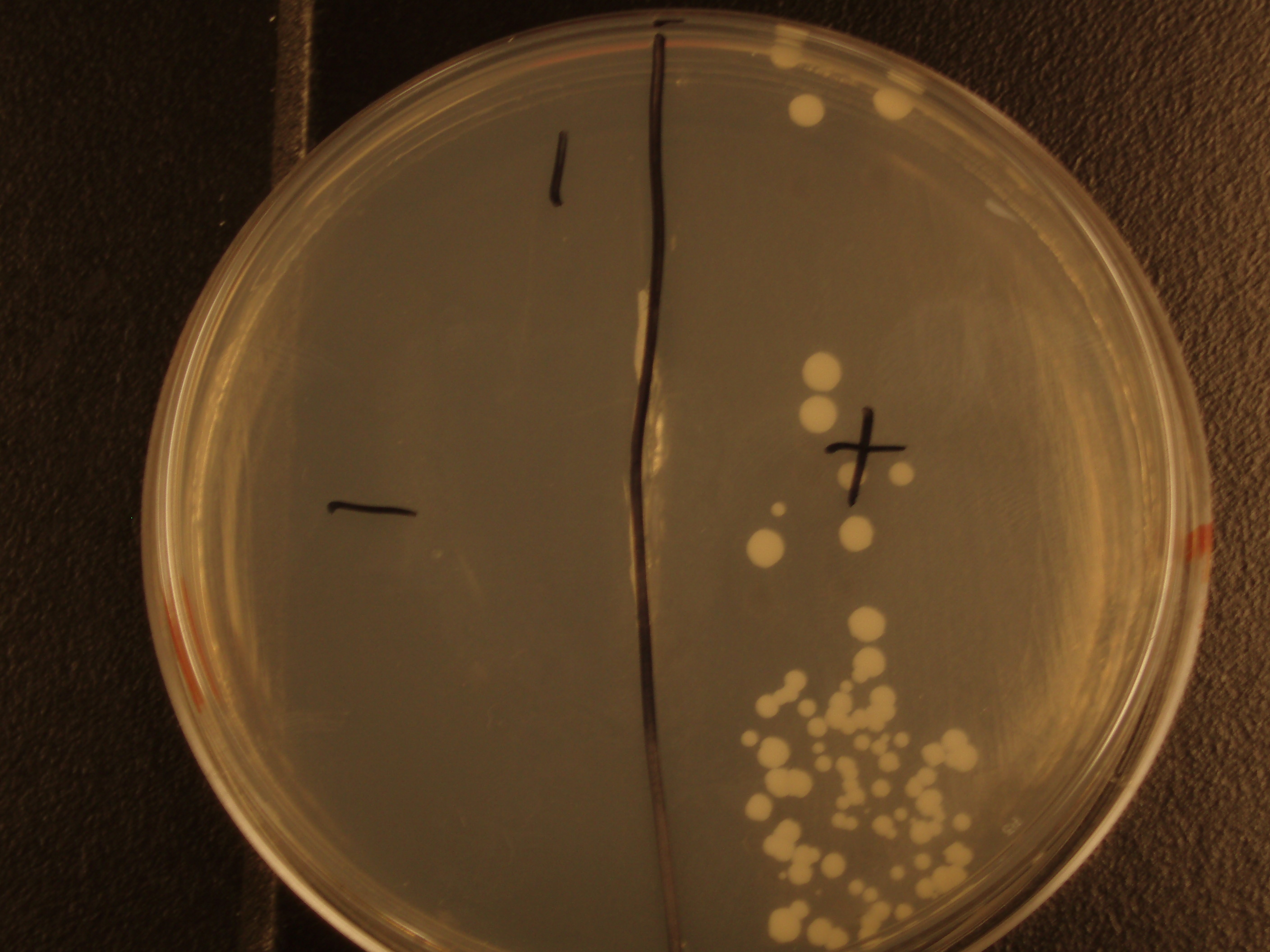
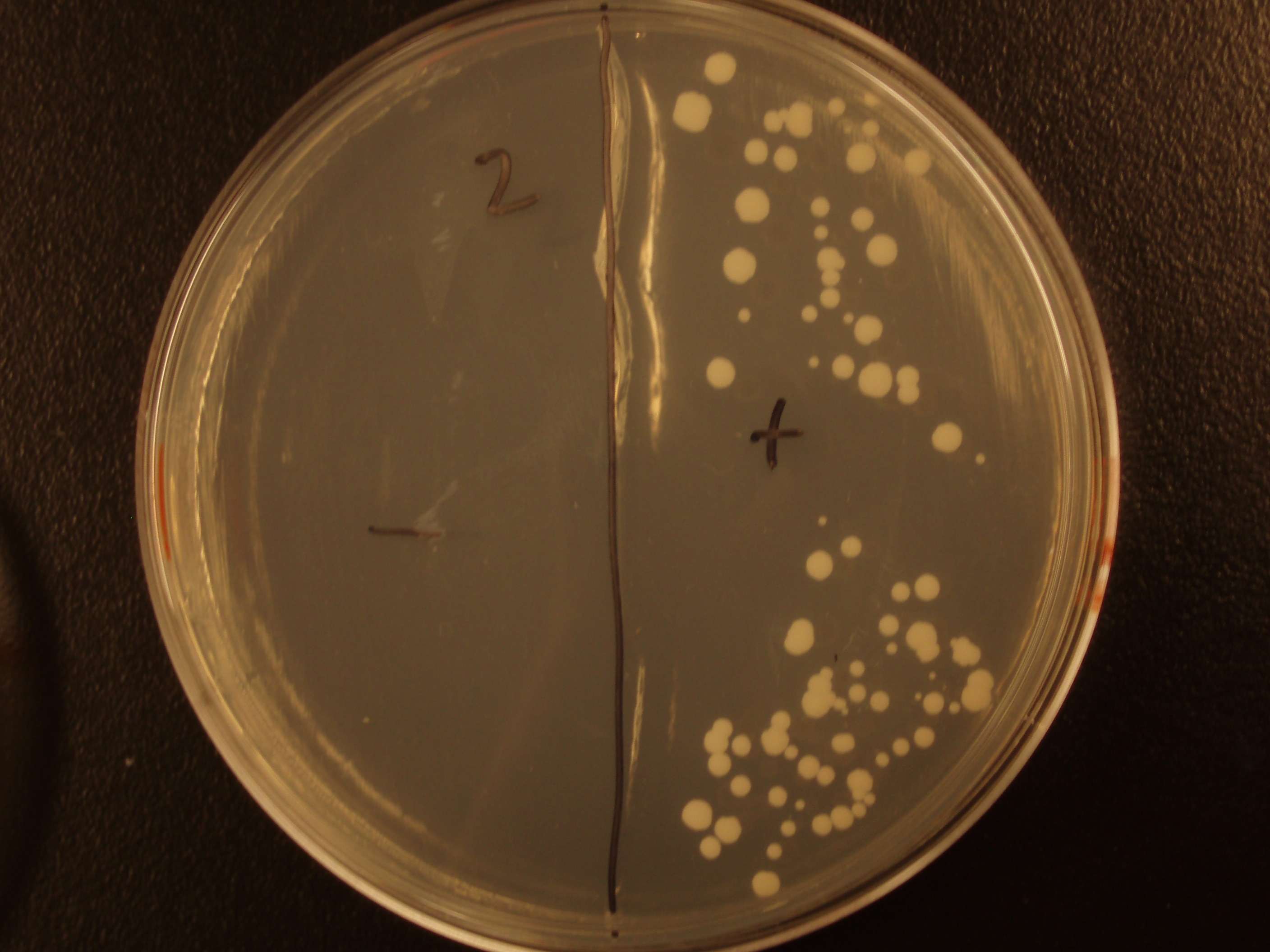
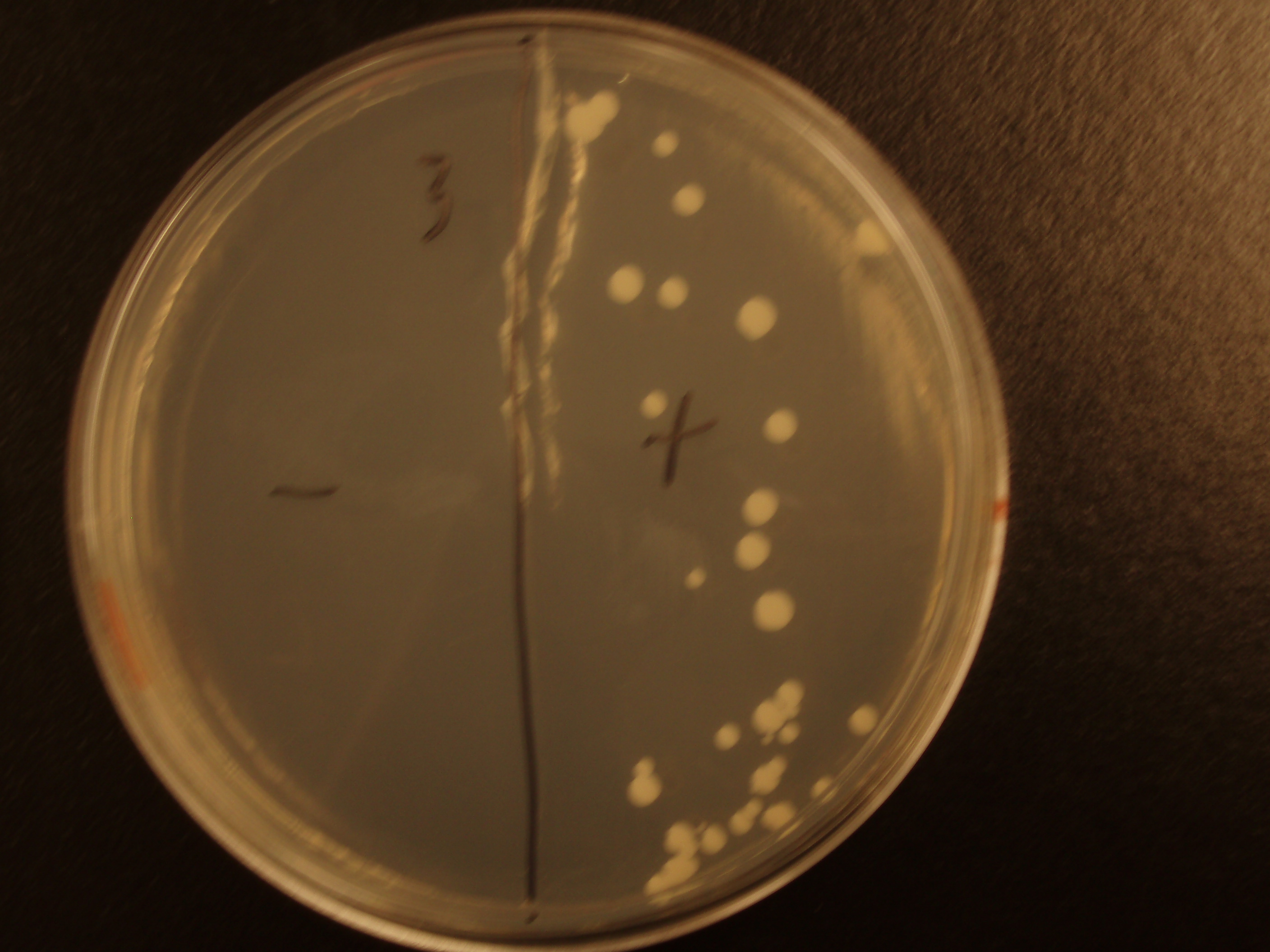
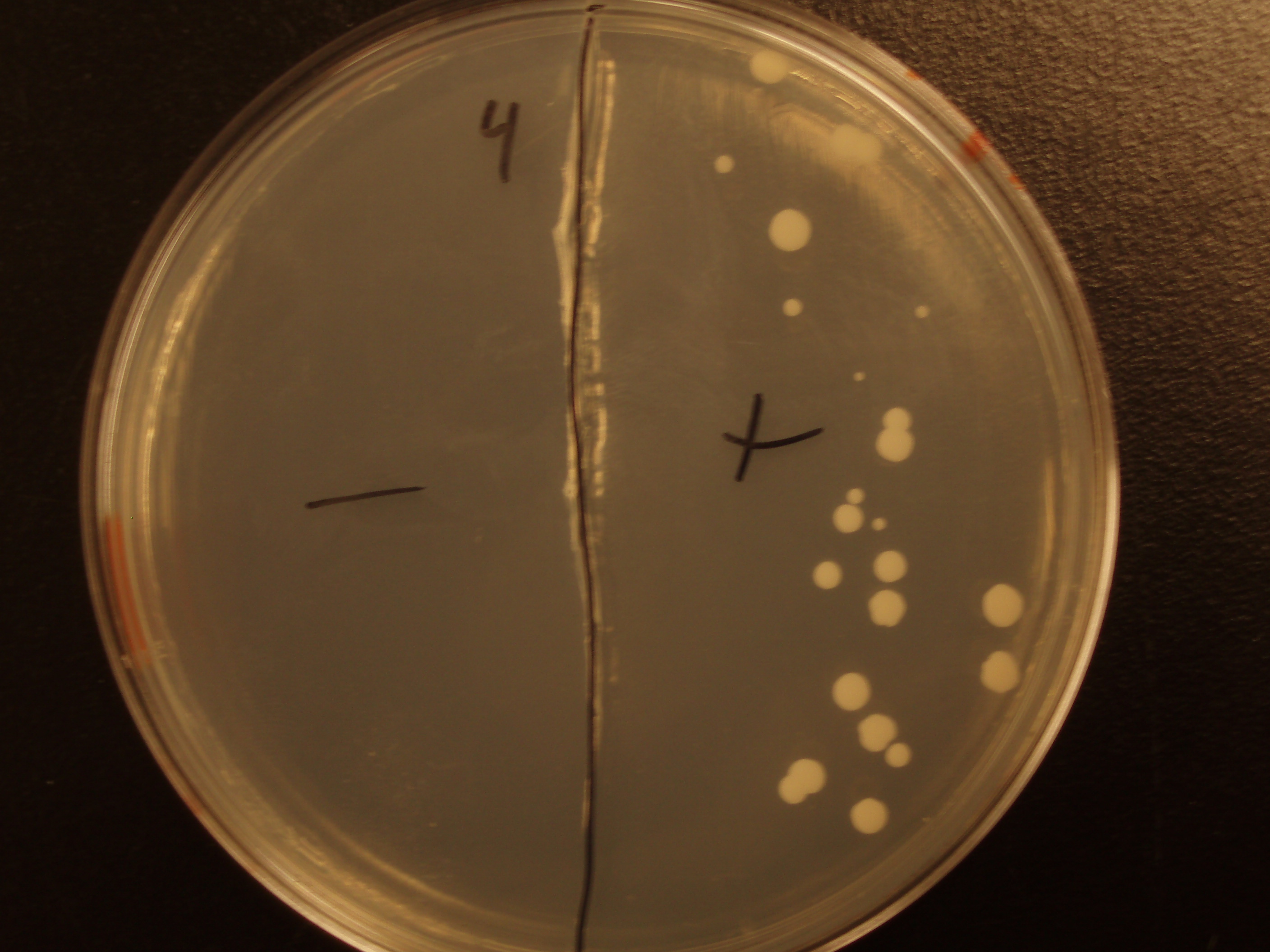
| Experiment # | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Bacteria (OD600@0.15 ≈ 3E7 bac/mL) | 250uL | 200uL | 100uL | 50uL |
| Yeast (OD600@0.1 ≈ 1E7 yeast/mL) | 250uL | 300uL | 400uL | 450uL |
| Yeast Colonies | 66 | 64 | 32 | 18 |
| Transconjugation Efficiency (Conjugation Events/Yeast Cell) | 2.64E-5 | 2.13E-5 | 8.00e-6 | 4.00e-6 |
| 
| 
| 
|
|---|
Yeast Shuttle Vector
One of the sub-projects involved in our controlled trans-kingdom conjugation project is the construction of a yeast shuttle vector. This plasmid will be stored and maintained in E. coli, and transferred to yeast via conjugation. Susbsequently, the plasmid will also be stored and maintained in yeast cells. Therefore, this yeast shuttle plasmid will have a bacterial origin of replication, a stable yeast origin of replication, a conjugative origin of transfer for RP4 mobilization, two genes which confer ampicilin and tetracycline resistance, a gene which imparts the ability to synthesize leucine, and finally the lac operon, which bestows the ability to digest lactose. A preliminary version of this plasmid, termed pAC88, has been created during prior research [2]. This pAC88 plasmid is a modified version of standard yeast plasmid YEp13; the origin of transfer site, OriT, from mobile plasmid RP4 was inserted into YEp13.
Functional pAC88 plasmids have been obtained from researchers at Leicester, which is a standard yeast cloning vector, YEp13, with an origin of transfer for RP4. The insertion of the RP4 OriT interrupts the gene for tetracycline resistance on YEp13, which is verifiable by transformed cells' sensitivity to tetracycline. In order to determine the efficacy of conjugation, S. cerevisiae was grown with E. coli in leucine-deficient media. Because pAC88 contains a gene which confers the ability to synthesize leucine, a strain of yeast which is normally auxotrophic for this amino acid was used. Successful conjugations resulted in yeast colonies that contained leucine.
There still remains the task of inserting the adaptive gene into the shuttle plasmid. Kluyveromyces lactis, a species of milk-yeast, posesses a region of chromosomal DNA that encodes for lactose digestion machinery. LAC4 and LAC12, two genes that encode for yeast galactosidase and lactose permease, respectively, are located near each other on the chromosome, and are divergently transcribed. Primers will be designed which are complementary to the transcriptional endpoints of LAC4 and LAC12, with hanging ends that correspond to the restriction enzyme HinDIII, an exploitable restriction site present in pAC88. A restriction digest on pAC88 and ligation with the LAC4-LAC12 cassette will insert the lactose digestion machinery into the yeast shuttle plasmid. By growing transformed yeast on a medium containing the molecular species X-gal, it will be determined whether the cassette has been successfully inserted if the yeast colonies appear blue.
[1] Sprague, George F., and Jack A. Heinemann. “Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast.” Nature. Institute of Molecular Biology. University of Oregon. 340. 20 Jul 1989.
[2] Cashmore, Annette M., Steven Bates, and Brian M. Wilkins. “IncP Plasmids Are Unusually Effective in Mediating Conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 Mating System.” Journal of Bacteriology. Dec 1998. pp 6538-6543.
 "
"