Team:Utah State/Project
From 2008.igem.org
(→Polyhydroxybutyrate) |
(→Methods) |
||
| Line 84: | Line 84: | ||
===Transformations=== | ===Transformations=== | ||
| + | A total of 26 transformations were successfully performed to become familiarized with the transformation process and to test different promoters and reporters. Three of these transformations were GPF plasmids with promoters of different strengths, taken from the iGEM promoter-testing kit. Another three of these transformations were BioBrick transformations. Top 10 competent E. coli cells were used for all transformations, excluding one transformation using a toxin-resistant strain. | ||
===Polymerase Chain Reaction=== | ===Polymerase Chain Reaction=== | ||
| Line 89: | Line 90: | ||
===PCR Primers=== | ===PCR Primers=== | ||
| + | Two sets of primers were created for PCR – those containing the prefix and suffix restriction sites and those without the prefix and suffix. The primers with the prefix and suffix already attached were designed to save subsequent ligations of the prefix and suffix from needing to be done. The primers without the prefix and suffix had greater affinity to the DNA template than those with them because of the absence of non-binding segments. | ||
===Vector Selection=== | ===Vector Selection=== | ||
| + | Two plasmids were primarily used during the course of the project – pSB1A3 and pSB3K3. pSB1A3 is an ampicillin-resistant plasmid, 2157 base pairs long, that was used as the vector for all submitted BioBricks. pSB3K3 is a Kanamycin-resistant plasmid, 2750 base pairs long, that was used for promoter testing in accordance with the iGEM promoter- testing protocol. | ||
=== Gel Electrophoresis === | === Gel Electrophoresis === | ||
Revision as of 01:50, 30 October 2008

|
| Home | The Team | The Project | Parts | Notebook | Protocols | Links |
|---|
Contents |
Abstract
The Utah State University iGEM team project is focused on creating an efficient system for producing and monitoring PHA production in microorganisms. One goal of our research is to develop and optimize a method, using fluorescent proteins, for the detection of maximum product yield of polyhydroxybutyrate (PHB, a bioplastic) in recombinant E. coli and in Cupriavidus necator. In order to develop an optimal PHB detection system, we worked to identify the most efficient reporter genes, and the best promoter sequences that would allow the GFP reporter to indicate maximum PHB production.
Project Objectives
DESIGN PHB reporter constructs (promoter region, reporter region, and PhaCAB cassette)
BUILD PHB reporter constructs
TEST for functionality of constructs
Introduction
Polyhydroxybutyrate
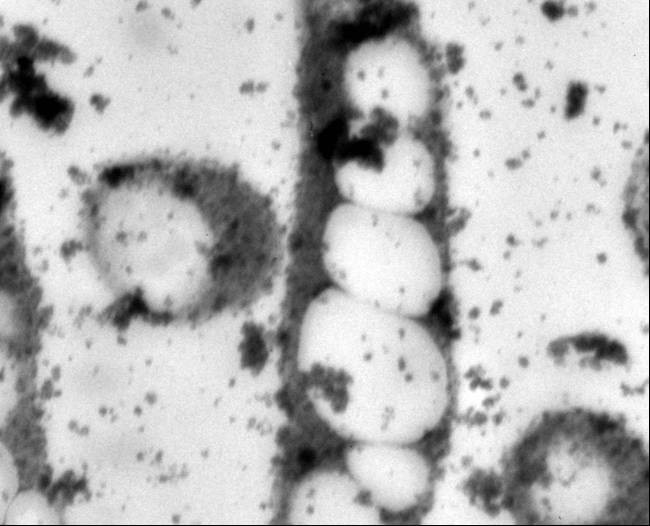
Concerns with the increasingly limited supply of fossil fuels and public anxiety over landfills oversaturated with non-biodegradable materials have also stimulated bioplastic research (Moran, 2006; Ojumu, 2004). The increasing cost and negative environmental effect of fossil hydrocarbon-derived conventional plastics have escalated the need for biodegradable alternatives from renewable sources. Polyhydroxyalkanoates are a class of over 80 polyesters synthesized by more than 300 microorganisms as a carbon and energy reserve. Specific types of PHAs exhibit material properties similar to those of petrochemical plastics (Lee, 1999; Lee, 1996). Polyhydroxybutyrate (PHB), the most prevalent PHA, has been of particular interest in commercial and medical applications because of its high tensile strength, biodegradability, and biocompatibility with human tissues (Lee, 1996b; Abe, 1992; Williams 2002). . PHA research has increased in an effort to understand its potential.
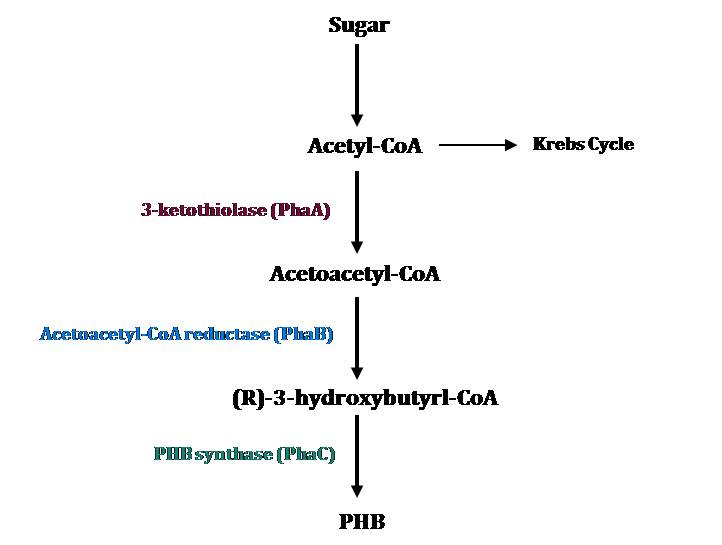
PHB Metabolic Pathways
The PHB cassette
Problems with PHB
1. COST
The current cost of industrial PHA production is estimated to be triple that of petrochemical plastics, which limits its widespread use.
2. PRODUCTION EFFICIENCY
Green Fluorescent Protein
GFP (Green Fluorescent Protein) is a protein originally isolated from the jellyfish Aequorea victoria, fluorescing when exposed to ultraviolet light. GFP is used as a tag, attached to proteins as a marker. Only those cells in which the tagged gene is expressed will fluoresce. In such a way GFP acts as a positive marker of tranformation. In our laboratory, GFP is used as an expression reporter of PHB. The GFP gene primarily used in this project was GFP E0240 BioBrick supplied by iGEM.
Methods
Organisms
Two organisms were used as genetic sources for PHB: Cupriavidus necator and an Escherichia coli strain containing the PHB operon in a pBluescript vector. The E. coli strain proved to be easier to use as a PCR template because of the ability to miniprep the plasmid DNA to use as more pure template.
Transformations
A total of 26 transformations were successfully performed to become familiarized with the transformation process and to test different promoters and reporters. Three of these transformations were GPF plasmids with promoters of different strengths, taken from the iGEM promoter-testing kit. Another three of these transformations were BioBrick transformations. Top 10 competent E. coli cells were used for all transformations, excluding one transformation using a toxin-resistant strain.
Polymerase Chain Reaction
Nine sequences were targeted out of the PHB operon for amplification. Five PCR targets were promoter sequences: 5’phaCproF/ATG R, 5’phaCproF/SD R, 5’phaCproF/TC R, -35F/ATG R, and -35F/TC R. Four PCR targets were from phaCAB gene complex: phaC, phaA, phaB, and phaCAB. A variety of annealing temperatures and Mg++ concentrations were tried, and it was found that 60˚C annealing temperature and 12% (v/v) Mg++ concentration were optimal. Two sets of primers were used: those without the BioBrick prefix and suffix attached and those with them already attached. The use of the PCR conditions just described enabled the use of the latter, saving subsequent ligations from needing to be done. Such PCR reactions were successful with all of the promoter regions and the phaB gene.
PCR Primers
Two sets of primers were created for PCR – those containing the prefix and suffix restriction sites and those without the prefix and suffix. The primers with the prefix and suffix already attached were designed to save subsequent ligations of the prefix and suffix from needing to be done. The primers without the prefix and suffix had greater affinity to the DNA template than those with them because of the absence of non-binding segments.
Vector Selection
Two plasmids were primarily used during the course of the project – pSB1A3 and pSB3K3. pSB1A3 is an ampicillin-resistant plasmid, 2157 base pairs long, that was used as the vector for all submitted BioBricks. pSB3K3 is a Kanamycin-resistant plasmid, 2750 base pairs long, that was used for promoter testing in accordance with the iGEM promoter- testing protocol.
Gel Electrophoresis
DNA gel electrophoresis, 1% w/v agarose, was used for analysis of PCR products, digested plasmid, and ligation constructs. The cross-linked matrix formed by agarose gel separates DNA by size when an electrical field is created across the gel. DNA migrates toward the positive electrical anode by electromotive force because of the natural negative charge of the DNA’s phosphate-sugar backbone. Ethidium bromide and Sybr Green dyes selectively bind DNA and were used in various experiments for band observation. Select PCR, plasmid, and ligation products were excised from agarose gels for purification and further processing.
GFP Correlation
After some deliberation, it was decided to use Green Fluorescent Protein as the PHB marker. Two methods were proposed to carry this through. The first method would involve ligating the GFP gene into the PHB operon, thereby allowing simultaneous transcription of the PHB and GFP genes, regulated by the PHB-induction machinery. The second method would involve separate PHB and GFP regions – either by bacteria containing two plasmids, one with the PHB operon and one with the GFP operon, or by using a plasmid containing both operons but in separate locations on the plasmid. In order to test for the most effective regions of the PHB promoter, five regions of the promoter were targeted for PCR (see PCR section). It was decided to then test these promoters using the iGEM promoter testing kit, testing for GFP expression in the pSB3K3 plasmid. A spectrofluorometer was used for fluorescence measurement.
PHB Promoter Testing
BioBrick Part Assembly
Results
- Obtained sources of PHB DNA
a. C. necator
b. DSMZ E. coli with pBluescript PHB plasmid
- Performed PCR to extract various PHB sequences
a. 5 promoter sequences
b. 4 CAB sequences
- Performed ligations and transformations to create BioBricks
BioBricks
5’phaCpro1 in pSB1A3 5’phaCpro3 in pSB1A3 -35/TC in psB1A3
PHB B into pSB1A3 (high PCR DNA concentrations) (awaiting ligation)
BioBricks in earlier stages of completion: 5’phaCpro2 in pSB1A3 -35Pro in pSB1A3 PHB A in pSB1A3 PHB C in pSB1A3 PHB CAB in pSB1A3 After promoter sequence testing, PHB CAB with promoter of choice and optimal reporter (likely GFP)
Conclusions
References
1. Doi Y, Kunioka M, Nakamura Y, Soga K. 1986. Nuclear magnetic resonance studies on poly(B-hydroxybutyrate) and a copolyester of B-hydroxybutyrate and B-hydroxyvalerate isolated from Alcaligenes eutrophus H16. Macromolecules. 19:2860-2864.
2. Endy D. 2005. Foundations for engineering biology. Nature. 438(7067):449-53. International Genetically Engineered Machines competition. 17 Oct 2008. 26 Jul 2008. <https://igem.org>.
3. Kang Z, Wang Q, and H Zhang. 2008. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Biotechnological Products and Process Engineering. 79:203-208.
4. Knight TF. 2003. Idempotent Vector Design for Standard Assembly of BioBricks. Tech. rep., MIT Synthetic Biology Working Group Technical Reports.
5. Registry of Standard Biological Parts. 17 Oct 2008. 26 Jul 2008 <http://partsregistry.org/Main_Page>.
6. Shetty RP, Endy D, Knight Jr TF. 2008. Engineering BioBrick vectors from BioBrick parts. Journal of Biological Engineering. 2:5.
7. Verlindin RAJ, Hill DJ, Kenward MA, Williams CD, and I Radecka. 2007. Bacterial synthesis of biodegradable Polyhydroxyalkanoates. Journal of Applied Microbiology 102:1437–1449.
 "
"