Team:Chiba/Calendar-Home/2 September 2008
From 2008.igem.org
(Difference between revisions)
(→Team:Input) |
(→Team:Input) |
||
| (6 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
==Laboratory work== | ==Laboratory work== | ||
===Team:Input=== | ===Team:Input=== | ||
| - | + | UV irradiation test | |
| - | Incubated cultures (from Glycerol Stocks) with 2ml of LB-Ampicillin Medium for 12 hours at 37 degrees. | + | #Incubated cultures (from Glycerol Stocks) with 2ml of LB-Ampicillin Medium for 12 hours at 37 degrees. |
| - | + | #Pre-incubated was plated so as to produce about 1000 colonies.(Ptet:1,Ptet-RFP:1,PrecA-RFP:3) | |
| - | + | #We put Negative Control of Prec-RFP in a dark place. | |
| - | + | #To 8 new Amp plates (2.5cm or 6.5cm apart from UV source for 30sec,1min, 30min, and 1h each) cultures grown at 37°C for 12h were transfered using nitrocellulose filters. | |
| - | + | #These are controls with Ptet and Ptet-RFP colonies. | |
| - | + | #PrecA-RFP plates were exposed to UV(254nm) at a distance 2.5cm or 6.5cm apart. | |
| - | + | #A Negative Control of Prec-RFP was placed in a dark place. | |
| + | #After UV exposure for 30sec, 1min, 30min, or 1h (each with either 2.5 or 6.5cm from the UV source or without light exposure), we transfered using nitrocellulose colonies from these plates to Amp control plates. | ||
| + | #We then tested the amount of time required for RFP expression after UV exposure. To do this, we scanned in the exposed plates after the various exposure times and observed the color change. | ||
| - | + | result | |
| - | + | We were not able to visually observe RFP fluorescence from plates containing PrecA-FRP. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===Team:Communication=== | ===Team:Communication=== | ||
Latest revision as of 03:10, 30 October 2008
1 September 2008 <|> 3 September 2008
Contents |
Laboratory work
Team:Input
UV irradiation test
- Incubated cultures (from Glycerol Stocks) with 2ml of LB-Ampicillin Medium for 12 hours at 37 degrees.
- Pre-incubated was plated so as to produce about 1000 colonies.(Ptet:1,Ptet-RFP:1,PrecA-RFP:3)
- We put Negative Control of Prec-RFP in a dark place.
- To 8 new Amp plates (2.5cm or 6.5cm apart from UV source for 30sec,1min, 30min, and 1h each) cultures grown at 37°C for 12h were transfered using nitrocellulose filters.
- These are controls with Ptet and Ptet-RFP colonies.
- PrecA-RFP plates were exposed to UV(254nm) at a distance 2.5cm or 6.5cm apart.
- A Negative Control of Prec-RFP was placed in a dark place.
- After UV exposure for 30sec, 1min, 30min, or 1h (each with either 2.5 or 6.5cm from the UV source or without light exposure), we transfered using nitrocellulose colonies from these plates to Amp control plates.
- We then tested the amount of time required for RFP expression after UV exposure. To do this, we scanned in the exposed plates after the various exposure times and observed the color change.
result
We were not able to visually observe RFP fluorescence from plates containing PrecA-FRP.
Team:Communication
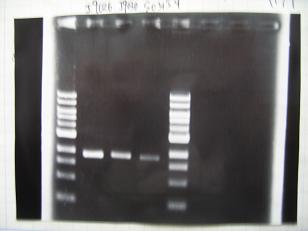
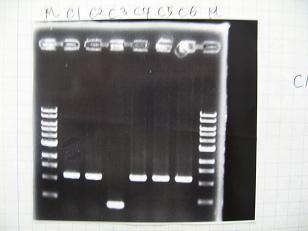
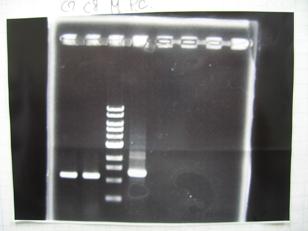
- (31/8)--->Gel Check
- (1/9)---> Colony PCR
- Colony PCR of 8 colonies from ligation plates (1/9:(1)[http://partsregistry.org/Part:BBa_K084009 BBa_K084009](R1~R8),(2)[http://partsregistry.org/Part:BBa_K084010 BBa_K084010](C1~C8)) and one from control plate([http://partsregistry.org/Part:BBa_F2620 BBa_F2620](2007)).
DNA Template 1 dNTP mix 5 Foward Primer 0.3 Reverse Primer 0.3 DNA polymerase TAQ 0.5 Thermopol Buffer 3 dH2O 20.5 TOTAL 30μL
- 95°C,5min -> ( 95°C,1min -> 52°C,1min -> 72°C,1min )・・・25cycles -> 72°C,10min -> 6°C
--->Gel CheckSample DNA 1 Loading Dye 1 dH2O 4 TOTAL 6μl - From left;
- Plac+RBS+RhlI+LVA
- R1 -> OK
- R2 -> Bad
- R3~R7 -> OK
- R8 -> Bad
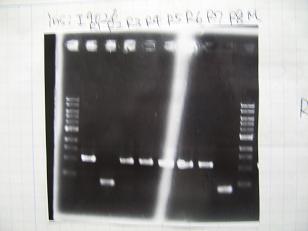
- From left;
- From left;
- Plac+RBS+CinI+LVA
- C1,C2 -> OK
- C3 -> Bad
- C4~C6 -> OK
- From left;
- Plac+RBS+CinI+LVA
- C7,C8 -> OK
- [http://partsregistry.org/Part:BBa_F2620 BBa_F2620](2007):Positive control -> OK
- --->(3/9)Mini prep
(1/9)--->Liquid Culture- Cultured the following cells (2mL LB-Amp, at 37°C, 7 hours)
- from transformed plates:
- [http://partsregistry.org/Part:BBa_K084007 BBa_K084007](Plac+RBS+LasI, Competent Cells : JW1908)
- [http://partsregistry.org/Part:BBa_K084008 BBa_K084008](Plac+RBS+RhlI, Competent Cells : JW1908)
- [http://partsregistry.org/Part:BBa_T9002 BBa_T9002](Ptet+RBS+LuxR+GFP, Competent Cells : JW1908)
- from Glycerol Stock:
- [http://partsregistry.org/Part:BBa_S03623 BBa_S03623](Ptet+RBS+LuxI, Competent Cells : JW1908)
- from transformed plates:
--->(3/9)Phenotype test
- Competent cells : XL10G 30μL
- [http://partsregistry.org/Part:BBa_C0161 BBa_C0161](2007)
- [http://partsregistry.org/Part:BBa_C0161 BBa_C0161](2006)
- [http://partsregistry.org/Part:BBa_C0261 BBa_C0261](2007)
- [http://partsregistry.org/Part:BBa_C0261 BBa_C0261](2006)
--->(4/9)Mini prep
Team:Output
Colony PCR
- [http://partsregistry.org/Part:BBa_R0010 BBa_R0010]+[http://partsregistry.org/Part:BBa_J52008 BBa_J52008]
Sample No. 1 culture 1 Fwd primer 1.5 Rev primer 1.5 Thermo pol Buffer 3 dNTP mix 3 Taq DNA pol (NEB) 0.2(1 unit) dH2O 19.8 TOTAL 30μl
-->95°C 5 min -->(95°C 1min -->50°C 30sec -->72°C 1min)x25 -->72°C 10min- [http://partsregistry.org/Part:BBa_R0079 BBa_R0079]
- [http://partsregistry.org/Part:BBa_R0071 BBa_R0071]
- [http://partsregistry.org/Part:BBa_R0077 BBa_R0077]
- [http://partsregistry.org/Part:BBa_R0078 BBa_R0078]
- [http://partsregistry.org/Part:BBa_R0062 BBa_R0062]
 "
"