Team:ETH Zurich/Wetlab/Overview
From 2008.igem.org
(→Outline) |
(→Outline) |
||
| (17 intermediate revisions not shown) | |||
| Line 34: | Line 34: | ||
===[[Team:ETH_Zurich/Wetlab/Switch_Circuit|Switch circuit]]=== | ===[[Team:ETH_Zurich/Wetlab/Switch_Circuit|Switch circuit]]=== | ||
| - | Expression of restriction enzymes that cut genomic DNA inside the cell is likely to decrease viability. Actually, the [[Team:Waterloo|Waterloo iGEM team]] is using restriction enzymes to kill | + | Expression of restriction enzymes that cut genomic DNA inside the cell is likely to decrease viability. Actually, the [[Team:Waterloo|Waterloo iGEM team]] is using restriction enzymes to kill cells in their project this year. Therefore, construction of a switch circuit, which allows to restrict expression of the restriction enzyme to a short period of time, is a crucial part of the project. The switch circuit allows expression of a gene under control of the lac repressor and rapid termination of lac-controlled expression despite presence of inducer by expression of an engineered LacI mutant, which is unresponsive to IPTG. |
| + | |||
| + | On the following pages, we will show a detailed description of how we are trying to achieve these three goals. | ||
== Outline == | == Outline == | ||
| Line 55: | Line 57: | ||
<br><br> | <br><br> | ||
'''Results:''' <br> | '''Results:''' <br> | ||
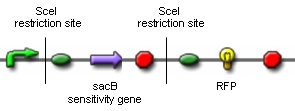
| - | We cloned a lac-inducible promoter in front of SceI and various inducible and constitutive promoters in front of the T4 ligase. During our attempts to design the construct coding for RFP flanked by SceI restriction sites, we managed to insert both restriction sites in front of and | + | We cloned a lac-inducible promoter in front of SceI and various inducible and constitutive promoters in front of the T4 ligase. During our attempts to design the construct coding for RFP flanked by SceI restriction sites, we managed to insert both restriction sites in front of and following the RFP by annealing oligonucleotide duplexes.<br><br> |
</div> | </div> | ||
| valign="top" align="center" width="450"| | | valign="top" align="center" width="450"| | ||
| Line 69: | Line 71: | ||
<br> | <br> | ||
'''Method:''' <br> | '''Method:''' <br> | ||
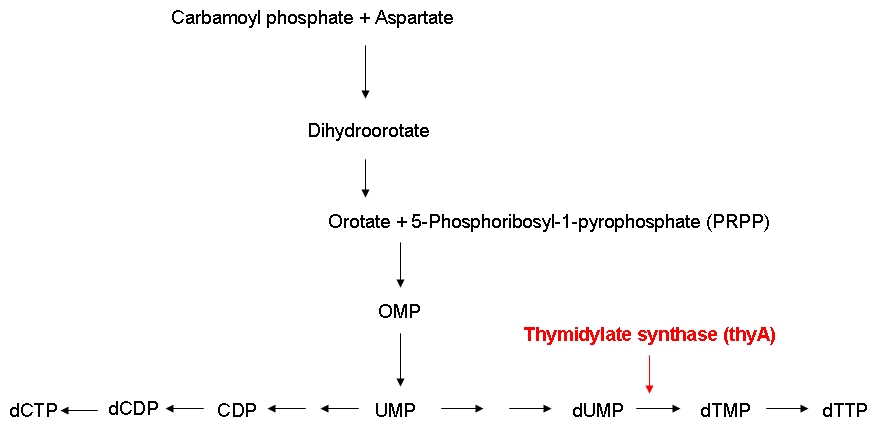
| - | In order to be able to examine the growth behaviors of ''E. coli'' strains carrying differently sized genomes, we ordered an E. coli strain (MDS42) whose genome had been reduced by 15 % using a targeted deletion approach. Also, we ordered the wild-type strain which the MDS42 | + | In order to be able to examine the growth behaviors of ''E. coli'' strains carrying differently sized genomes, we ordered an ''E. coli'' strain (MDS42) whose genome had been reduced by 15 % using a targeted deletion approach. Also, we ordered the wild-type strain which the MDS42 had been derived from. In order to be able to keep track of the individual growth rates of the wild-type and MDS42 strains if grown in a mixed culture, we wanted to label these two strains. Finally, for knocking out the thymidylate synthase, we decided to use phage transduction.<br><br> |
'''Results:''' <br> | '''Results:''' <br> | ||
| - | We successfully knocked out the thymidylate synthase, both in the wild-type and the MDS42 ''E. coli'' strains. Also | + | We successfully knocked out the thymidylate synthase, both in the wild-type and the MDS42 ''E. coli'' strains. Also, we managed to label the wild-type and the MDS42 ''E. coli'' strains by transformation of low-copy plasmids encoding different reporter proteins. Finally, we successfully performed growth experiments showing that the growth rate of thymidylate synthase knockout strains can be influenced by regulating the external thymidine supply.<br><br> |
</div> | </div> | ||
|- | |- | ||
| Line 77: | Line 79: | ||
'''3)''' [[Team:ETH_Zurich/Wetlab/Switch_Circuit|'''Switch circuit''']] | '''3)''' [[Team:ETH_Zurich/Wetlab/Switch_Circuit|'''Switch circuit''']] | ||
| - | + | [[Image:jr_pulsegen_5.jpg|center|300px|]] | |
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
'''Questions:''' <br> | '''Questions:''' <br> | ||
| - | * | + | * Can we construct a simple and at the same time robust pulse generator that allows pulses of variable duration without modification of the construct? |
| - | * | + | * Can we build a pulse generator that would work inside the chemostat? |
<br> | <br> | ||
'''Method:''' <br> | '''Method:''' <br> | ||
| - | We decided to | + | We decided to build a simple pulse generator based on LacI IS mutants, which upon expression rapidly silence lac-controlled expression despite presence of IPTG. This allows us to induce expression with IPTG and terminate expression by addition of tetracyline. Since it is independent of removal of inducer by complete exchange of medium, it can be used inside the chemostat.<br><br> |
'''Results:''' <br> | '''Results:''' <br> | ||
| - | + | Using PCR-based site-directed mutagenesis we generated eight different mutants of LacI and characterized them in a simple genetic experiment. As it turned out, all of them repressed lac-controlled expression even at high concentrations of IPTG. We constructed a pulse generator consisting of a tet-controlled LacI IS generator and a constitutive TetR expression cassette and demonstrated that leaky expression of the protein of interest as well as repression of lac-controlled expression due to leaky expression of LacI IS are not problematic. <br><br> | |
| - | <br><br> | + | |
</div> | </div> | ||
| | | | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Latest revision as of 03:34, 30 October 2008
|
OverviewIn order to approach our goal of creating an E. coli strain carrying a minimal genome, there are three main problems that have to be overcome:
Chemostat selection: introduce a limitation that confers a growth advantage to organisms with smaller genomes Switch circuit: design a biobrick that provides for short-term synthesis of the desired gene products
Genome ReductionTo prove that in vivo restriction and religation is possible is fundamental to our project which relies on short-term expression of a restriction enzyme and a ligase. While the restriction enzyme will randomly cut DNA, the simultaneous or shortly delayed synthesis of the ligase should religate the DNA. If the DNA is cut at several sites, religation will lead to exclusion of chromosomal fragments in a random manner. Chemostat selectionIn the continuous culture of a chemostat, those organisms with the highest rate of proliferation will overgrow those with a smaller growth rate. In order to bypass the need of selecting for those E. coli which have successfully reduced their genomes by massive screening of thousands of clones, we need to introduce a constraint that confers a growth advantage to organisms with smaller genomes. We have chosen to introduce mutations in the nucleotide synthesis pathway to achieve this goal. This will render DNA replication the rate-limiting step of proliferation and therefore be advantageous to organisms with small genomes. Switch circuitExpression of restriction enzymes that cut genomic DNA inside the cell is likely to decrease viability. Actually, the Waterloo iGEM team is using restriction enzymes to kill cells in their project this year. Therefore, construction of a switch circuit, which allows to restrict expression of the restriction enzyme to a short period of time, is a crucial part of the project. The switch circuit allows expression of a gene under control of the lac repressor and rapid termination of lac-controlled expression despite presence of inducer by expression of an engineered LacI mutant, which is unresponsive to IPTG.
Outline
|
 "
"