Team:MIT
From 2008.igem.org
(→Models) |
|||
| (69 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<!--- The Mission, Experiments ---> | <!--- The Mission, Experiments ---> | ||
| - | {| style="color:#1b2c8a;background-color:# | + | [[Image:MitBanner.jpg|center]] |
| + | |||
| + | |||
| + | {| style="color:#1b2c8a;background-color:#CCCCCC;" cellpadding="3" cellspacing="1" border="1" bordercolor="#green" width="62%" align="center" | ||
!align="center"|[[Team:MIT|Home]] | !align="center"|[[Team:MIT|Home]] | ||
!align="center"|[[Team:MIT/Team|The Team]] | !align="center"|[[Team:MIT/Team|The Team]] | ||
!align="center"|[[Team:MIT/Project|The Project]] | !align="center"|[[Team:MIT/Project|The Project]] | ||
| + | !align="center"|[[Team:MIT/Experiments|Experiments]] | ||
!align="center"|[[Team:MIT/Parts|Parts Submitted to the Registry]] | !align="center"|[[Team:MIT/Parts|Parts Submitted to the Registry]] | ||
| - | !align="center"|[[Team:MIT/Modeling| | + | !align="center"|[[Team:MIT/Modeling|Results]] |
!align="center"|[[Team:MIT/Notebook|Notebook]] | !align="center"|[[Team:MIT/Notebook|Notebook]] | ||
|} | |} | ||
| + | |||
<hr class=divider> | <hr class=divider> | ||
<div id="about"> | <div id="about"> | ||
| - | |||
| - | |||
| - | + | ==Biogurt: A Sustainable and Savory Drug Delivery System== | |
| - | + | ''' | |
| - | + | Streptococcus mutans, the main cause of dental caries, binds to glycoproteins on the teeth. A clinical study (Kelly CG et al.; Nature Biotechnol. 1999) isolated the 20aa functional segment (p1025) that S.mutans uses to attach to the teeth. p1025 competitively inhibits the binding of S.mutans, causing unharmful bacteria to grow in its place, preventing the recolonization of S.mutans for 90 days. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We are engineering Lactobacillus bulgaricus, a bacteria commonly found in yogurt, to produce and secrete this peptide under a promoter activated by lactose. | |
| - | + | ||
| - | + | The peptide p1025 could simply be added to any food. But production of this peptide by L. bulgaricus is an independent process, so inserting the gene into live bacteria in yogurt will enable continuous production. Since a new batch of yogurt can be made using the bacteria from a small amount of the old batch, a continuous supply of teeth-cleaning yogurt will be available from the first successfully engineered batch. This could be the key to providing effective dental health care in underdeveloped rural communities, especially if yogurt is already an integral part of the diet. | |
| - | + | ||
| - | + | Also, the p1025 gene could be replaced by any other gene, so this same expression system could be used to produce other useful peptides. Yogurt with modified bacteria will provide a cheap, efficient, and delicious way to distribute vitamins, vaccines and more. | |
| - | + | ''' | |
| - | + | ||
| - | // | + | ==Results== |
| - | + | ====Characterization==== | |
| - | + | *plasmid pTG262 - Last year's Edinburgh team worked with the plasmid pTG262 in trying to transform it into different gram-positive bacteria. They had not transformed it into ''Lactobacillus'' but were optimistic that it could be, since pTG262 is known to be able to replicate in strains of ''Lactobacillus''. Dr. Chris French was very helpful and graciously supplied us with this plasmid. Working extensively with this plasmid with all of our strains of bacteria, we were unable to successfully transform pTG262 into ''Lactobacillus'', and we have concluded that it is our opinion that pTG262 '''cannot''' be electroporated into ''Lactobacillus delbruckii''. We have added '''characterization''' to this part's main page and user review page, [http://partsregistry.org/wiki/index.php?title=Part:BBa_I742103 '''here''']. | |
| - | + | ||
| - | + | * p1025-This sequence codes for a short peptide, found to competitively inhibit binding of S.mutans to the tooth surface (CG et al.; Nature Biotechnol. 1999). S.mutans takes in sugars and secretes lactic acid, causing dental cavities, so introduction of this peptide into the mouth prevents colonies. This part uses the modified Silver BioBrick prefix and suffix to allow for protein construction. | |
| - | == | + | |
| - | ''' Our | + | ====Models==== |
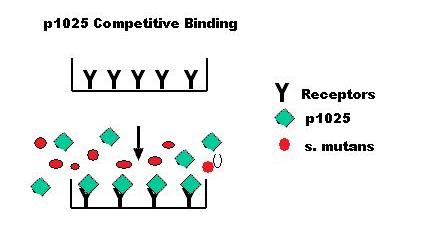
| + | [[Team:MIT/Competitive Binding Model|Competitive Binding Model]] | ||
| + | [[Image:Competitve binding.jpg]] | ||
| + | *This model shows that p1025 inhibits binding of s. mutans to the salivary receptors on the tooth surface. p1025 serves as the competitive peptide inhibitor of bacterial adhesion in this model. Our team is the first to not only invent a tooth binding assay but characterize p1025 as a competitive inhibitor. | ||
| + | |||
| + | ====New Methods==== | ||
| + | *''S.mutans'' Binding Assay | ||
| + | *p1025 Competitive Binding Assay | ||
| + | * [[Team:MIT/Tooth_binding_assay_protocol|Novel Tooth Binding Assay]] | ||
| + | * [[Team:MIT/Transforming_Lactococcus lactis|''Lactococcus lactis'' Transformation Protocol]] | ||
| + | * [http://openwetware.org/wiki/L._acidophilus_transformation ''Lactobacillus acidophilus'' Transformation Protocol] | ||
| + | * [http://openwetware.org/wiki/Lactobacillus_transformation ''Lactobacillus delbruckii'' Transformation Protocol] | ||
| + | * [http://openwetware.org/wiki/Lactobacillus_culture Culturing ''Lactobacillus''] | ||
| + | * [http://openwetware.org/wiki/Lactobacillus_miniprep ''Lactobacillus delbruckii'' Miniprep protocol''] | ||
| + | * [http://openwetware.org/wiki/Lactobacillus_planarum_miniprep ''Lactobacillus plantarum'' Miniprep protocol''] | ||
| + | |||
| + | ====Lactobacillus Work==== | ||
| + | *We '''successfully transformed Lactobacillus delbruckii subsp. bulgaricus and Lactobacillus delbruckii subsp. lactis''' with a modified version of Serror et al's electrotransformation procedure. Our [http://openwetware.org/wiki/Lactobacillus_transformation '''transformation protocols'''] are found here. We also plan to submit these plasmids, with biobrick sites inserted in them, to the registry. These plasmids are [http://partsregistry.org/wiki/index.php?title=Part:BBa_K128008 '''pJK650'''] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K128007 '''pLEM415''']. | ||
| + | |||
| + | *We are currently working on a method to effectively miniprep plasmid DNA from our ''Lactobacillus''. For information about our protocol for doing so, please click [http://openwetware.org/wiki/Lactobacillus_miniprep '''here''']. | ||
| + | |||
| + | *We have built an expression system for Lactobacillus to secrete protein, including individual biobricked parts of a Lactobacillus [http://partsregistry.org/wiki/index.php?title=Part:BBa_K128006 '''promoter'''] and a Lactobacillus [http://partsregistry.org/Part:BBa_K128004 '''signal sequence''']. | ||
| + | |||
| + | *Please visit the results page and experiments page of this wiki for full report of all our results and team-generated protocols!!!! | ||
| + | |||
| + | ==Acknowledgements== | ||
| + | =====Graduate Advisers===== | ||
| + | Scott Carlson, Felix Moser, Chia-Yung Wu, Lav Varshney, Vikramaditya Yadav, Woo Chung, Rachel Hilmer, Robbier Barbero, Brian Cook, Jyoti Goda, Laure-Anne Ventouras | ||
| + | |||
| + | =====Faculty Advisors:===== | ||
| + | Drew Endy, Tom Knight | ||
| - | + | =====Others===== | |
| - | + | Isadora Deese, | |
| + | Dr. Pascale Serror at L'Institut National de la Recherche Agronomique, | ||
| + | Dr. Bernhard Heinrich at the University of Kaiserslautern, | ||
| + | Dr. Chris French at the University of Edinburgh, | ||
| + | Dr. Daniel Smith at the Forsyth Institute, | ||
| + | Joey Davis at the Sauer Lab at MIT, | ||
| + | The Fink Lab at the Whitehead Institute, | ||
| + | Emiko Bare at the Keating Lab at MIT, | ||
| + | Jennifer Hou at the Baker Lab at MIT, | ||
| + | The UROP office and Biological Engineering Department for financial support | ||
|- | |- | ||
| Line 55: | Line 92: | ||
<!--- The Mission, Experiments ---> | <!--- The Mission, Experiments ---> | ||
| - | {| style="color:#1b2c8a;background-color:# | + | {| style="color:#1b2c8a;background-color:#CCCCCC;" cellpadding="3" cellspacing="1" border="1" bordercolor="#green" width="62%" align="center" |
!align="center"|[[Team:MIT|Home]] | !align="center"|[[Team:MIT|Home]] | ||
!align="center"|[[Team:MIT/Team|The Team]] | !align="center"|[[Team:MIT/Team|The Team]] | ||
!align="center"|[[Team:MIT/Project|The Project]] | !align="center"|[[Team:MIT/Project|The Project]] | ||
| + | !align="center"|[[Team:MIT/Experiments|Experiments]] | ||
!align="center"|[[Team:MIT/Parts|Parts Submitted to the Registry]] | !align="center"|[[Team:MIT/Parts|Parts Submitted to the Registry]] | ||
!align="center"|[[Team:MIT/Modeling|Modeling]] | !align="center"|[[Team:MIT/Modeling|Modeling]] | ||
!align="center"|[[Team:MIT/Notebook|Notebook]] | !align="center"|[[Team:MIT/Notebook|Notebook]] | ||
|} | |} | ||
Latest revision as of 09:46, 30 October 2008
Biogurt: A Sustainable and Savory Drug Delivery SystemStreptococcus mutans, the main cause of dental caries, binds to glycoproteins on the teeth. A clinical study (Kelly CG et al.; Nature Biotechnol. 1999) isolated the 20aa functional segment (p1025) that S.mutans uses to attach to the teeth. p1025 competitively inhibits the binding of S.mutans, causing unharmful bacteria to grow in its place, preventing the recolonization of S.mutans for 90 days. We are engineering Lactobacillus bulgaricus, a bacteria commonly found in yogurt, to produce and secrete this peptide under a promoter activated by lactose. The peptide p1025 could simply be added to any food. But production of this peptide by L. bulgaricus is an independent process, so inserting the gene into live bacteria in yogurt will enable continuous production. Since a new batch of yogurt can be made using the bacteria from a small amount of the old batch, a continuous supply of teeth-cleaning yogurt will be available from the first successfully engineered batch. This could be the key to providing effective dental health care in underdeveloped rural communities, especially if yogurt is already an integral part of the diet. Also, the p1025 gene could be replaced by any other gene, so this same expression system could be used to produce other useful peptides. Yogurt with modified bacteria will provide a cheap, efficient, and delicious way to distribute vitamins, vaccines and more. ResultsCharacterization
Models
New Methods
Lactobacillus Work
AcknowledgementsGraduate AdvisersScott Carlson, Felix Moser, Chia-Yung Wu, Lav Varshney, Vikramaditya Yadav, Woo Chung, Rachel Hilmer, Robbier Barbero, Brian Cook, Jyoti Goda, Laure-Anne Ventouras Faculty Advisors:Drew Endy, Tom Knight OthersIsadora Deese, Dr. Pascale Serror at L'Institut National de la Recherche Agronomique, Dr. Bernhard Heinrich at the University of Kaiserslautern, Dr. Chris French at the University of Edinburgh, Dr. Daniel Smith at the Forsyth Institute, Joey Davis at the Sauer Lab at MIT, The Fink Lab at the Whitehead Institute, Emiko Bare at the Keating Lab at MIT, Jennifer Hou at the Baker Lab at MIT, The UROP office and Biological Engineering Department for financial support | ||||||||
|
| ||||||||
| Home | The Team | The Project | Experiments | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"