Team:UC Berkeley Tools
From 2008.igem.org
(Prototype team page) |
|||
| (296 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <!-- | + | <font size="4"><center>Winner of the "Best Software Tool" award, iGEM 2008! (see the winning presentation [https://2008.igem.org/files/video/UC_Berkeley_Tools.mp4 here])</center></font> |
| + | [[Image:Clotho_Title_small.png|center]] | ||
| + | <!--- <center>[http://biocad-server.eecs.berkeley.edu/wiki/index.php/Clotho_Development#Downloads_and_Information <b>The Alpha release is here.</b>]</center> ---> | ||
| + | {{Template:ClothoMenu}} | ||
| + | __NOTOC__ | ||
| + | |||
| + | <!--- [[Image:DayShot_Campanile.png|right|125 px]] ---> | ||
| + | <!--- [[Image:stacked_screenshots.png|thumb|225 px|right|Clotho Screenshots]] ---> | ||
<html> | <html> | ||
| - | < | + | <body> |
| - | < | + | <table width="950" border="0" bgcolor=#000080> |
| - | + | <tr> | |
| + | <td><p> | ||
| + | |||
| + | <font size="6">W</font>ith >700 available parts and >2000 defined parts in the <A href="http://partsregistry.org/Main_Page"> MIT Registry of Standard Biological Parts</A>, additional part repositories being developed, and countless more parts local to the lab, synthetic biologists are now faced with the challenge of organizing, sorting, and editing these parts from an ever-growing collection of DNA. Computational tools are becoming increasingly needed to assist biologists to do these jobs, and it is no secret that many tools are being introduced into the community to help serve these needs. It is important that new tool developments in this area remain both firmly rooted in the realities of the present while being forward thinking and flexible.</p> | ||
| + | |||
| + | <p><font size="6">C</font>lotho is a toolset designed in a <a href="http://www.gigascale.org/pubs/141.html"> platform-based design </a> paradigm to consolidate all these tools into one working, integrated toolbox. Clotho is based on a <a href = "https://2008.igem.org/Team:UC_Berkeley_Tools/Project#Project_Architecture"> core-and-hub system </a> which manages multiple connections, each connection performing tasks in some self-sufficient manner while connected to all other connections via the core. Examples of connections include sequence viewers/editors, parts database managers, assembly algorithm GUIs, and much more. Computer-savvy biologists can create new connections which will then be integrated into Clotho. These pieces can be kept locally in ones lab or made public and shared with the community. In this tool-consolidation and customization it is hoped that, in the future, biologists need not look further for a program that provides the answer to their ever growing design needs.</p> | ||
| + | |||
| + | </td> | ||
| + | <td> | ||
| + | <div align="right" class="youtube"> | ||
| + | <object width="425" height="350"> <param name="movie" value="http://www.youtube.com/v/_wD-DgnopVU"> </param> <embed src="http://www.youtube.com/v/_wD-DgnopVU" type="application/x-shockwave-flash" width="425" height="350"> </embed> </object> | ||
</div> | </div> | ||
| - | < | + | </td> |
| - | + | </tr> | |
| - | </ | + | </table> |
| - | < | + | </body> |
| - | + | </html> | |
| - | + | <!--- The Mission, Experiments ---> | |
| - | < | + | <!-- Other images include: DOP_DougPic.png, AnneAtTheDOP.png--> |
| - | + | [[Image:side_by_side_screenshots.jpg|thumb|400 px|Clotho Screenshots]] | |
| + | {{Template:ClothoDownloadButton}} | ||
| + | <br clear="all"> | ||
| + | ==Clotho Overview== | ||
| - | < | + | <center>{{Template:ClothoDesignFlow}}</center> |
| + | <html><font size="6">A</font></html>mong many possible top-level explanations of Clotho, one of the simplest is the "Clotho Data Structure"-centric view. In this view, biological part data is passed back and forth between Clotho components and to files and databases outside Clotho. Some of the possible flows of data are represented by the slew of arrows in the diagram above. | ||
| - | + | <big><b>At the Center: Clotho Data Structure</b></big> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Local and standard databases vary from one lab to another, and the way to grab biological part data from one database may require a completely different route than another. The [[Team:UC_Berkeley_Tools/Project#6._Clotho_Data_Structure|'''Clotho Data Structure''']], in essence, can be viewed as the Rosetta stone of Clotho's environment. In a nutshell, it allows any developer to grab relevant information about a part from any given database. This is made possible when a developer "binds" fields in a database to specific Clotho Data Structure objects which have a specific semantic associated with them. When a user wants to grab the sequence belonging to a particular Biobrick, for example, the Clotho Data Structure, once "bound" to a particular database, will be able to grab the sequence from the database and give it to Clotho. Want to know who owned the vector from jca1021 in database A and send that information to database B? Easy. Bind the data structure with both databases, grab the data from database A, and tell Clotho where in database B to place the data. This binding not only gives unstandardized data meaning, it also provides a common language by which Clotho developers can communicate. | |
| - | + | ||
| - | + | <big><b>Views/Functions</b></big> | |
| - | + | ||
| - | + | Each category above merits its own explanation, as each is vitally important to the project. Perhaps a good place to start are the '''views/functions'''. Here users are able to see and manipulate data, such as part information or the nucleotide sequence. Examples include the [[Team:UC_Berkeley_Tools/Project#Sequence_View|sequence view]] or the [[Team:UC_Berkeley_Tools/Project#Plate_Manager|plate manager]]. | |
| - | + | ||
| - | + | ||
| - | ! | + | <big><b>Interfaces</b></big> |
| - | + | ||
| - | + | Not all views/functions are self-sufficient, and so another set of components, called '''interfaces''', are involved in assisting these views/functions for functionality. Take for example the [[Team:UC_Berkeley_Tools/Project#Sequence_View|sequence view]] (a view many ApE lovers would recognize). How would the sequence view know how to highlight features or enzymes if Clotho didn't know what features or enzymes to look for? That's where the [[Team:UC_Berkeley_Tools/Project#Feature_Library_Collection|features library]] and [[Team:UC_Berkeley_Tools/Project#Enzyme_Library|enzyme library]] come in. In this particular example, the two interfaces store and manage information that the sequence view will have to frequent in order to perform vital operations. Other interfaces include a [[Team:UC_Berkeley_Tools/Project#Parts_Manager|parts manager]] and [[Team:UC_Berkeley_Tools/Project#Database_Manager|database manager]]. These allow the user to view part data and relationships. Not only can the source of parts be viewed but users can maintain personal collections of parts as well. | |
| + | |||
| + | |||
| + | <big><b>Algorithm Environment</b></big> | ||
| + | |||
| + | But what if you want to do ''more'' with your data you've manipulated? What if, after gathering all your parts, you want to calculate the optimal assembly scheme for a particular goal composite part? This is where the [[Team:UC_Berkeley_Tools/Project#Algorithm_Manager|'''algorithm environment''']] kicks in. The algorithm environment allows developers to make systems that take both quick one-liner inputs or more complex input from the Clotho Data Structure and spit out one-liner or more sophisticated outputs. The [[Team:UC_Berkeley_Tools/Project#Algorithm_Manager|algorithm manager]] allows not only that both approaches can be performed but each are recognized seamlessly. Let's say you wanted to program a partial matching utility into Clotho, where two sequences are given as inputs and a streamlined one-line primer match were spit back out. Algorithm manager provides you all the basic facilities to code such a utility. | ||
| + | |||
| + | |||
| + | <big><b>Clotho Connectors</b></big> | ||
| + | |||
| + | Given all these features, there still need to be components that allow Clotho to connect to the outside world. [[Team:UC_Berkeley_Tools/Project#mySQL_Configuration_Manager|'''Clotho connectors''']], so aptly named, perform this exact purpose. These components allow users to connect to databases through internet connections, or import and export data from existing files (think .gb, .ape, .str, etc). Connectors can also translate the results of Clotho back into any number of formats if the user so wishes. | ||
| + | |||
| + | |||
| + | <big><b>Clotho Plug-in Environment</b></big> | ||
| + | |||
| + | But last, and certainly not least, is the [[Team:UC_Berkeley_Tools/Project#7._Clotho_Plug-In_System|'''Clotho plug-in environment''']]. This powerful environment allows developers both associated and not associated with the Clotho Project to create new and customized plug-ins to integrate with their versions of Clotho. Clotho was designed to be as ''fluid'' and ''flexible'' as possible! While the project is indeed open source, and users can always make new components by making new connections (see [[Team:UC_Berkeley_Tools/Project#The_Project|Project Technical]]), developers can now program personalized tools without knowing anything about the Clotho architecture, save for the plug-in environment! Plug-ins are a bit of an intermediate between a Clotho algorithm and a Clotho connection. This allows for relative customization, but also makes the programmer's life easier. They are rooted in the [http://jpf.sourceforge.net/ Java Plugin Framework (JPF)] and should provide a powerful, community driven aspect to Clotho. | ||
| + | |||
| + | |||
| + | We have spent long and hard working hours to develop this tool, and we sure hope you enjoy it. Play around with it and be sure to let us know what you think! | ||
| + | |||
| + | <big><center>'''WELCOME TO CLOTHO!'''</center></big> | ||
| + | <center>''Nade, Matt, Anne, and Doug''</center> | ||
| + | |||
| + | <!---The Clotho design environment is centered around the ''Clotho Data Structure''. This object unifies information about a design in a standard way. This links various pieces of data related in a design to a powerful operational semantic. This semantic allows for design information to be related to databases containing part information, infer information about parts in a design, perform part look-ups in real time, and apply algorithms to designs. By taking potentially generic external information and bringing it into the Clotho Data Structure, non-standard designs can become standardized and then disseminated to a wider design community. | ||
| + | |||
| + | Shown in the figure are the major components in the Clotho design environment. In the center of the diagram is the Clotho Data structure...yada yada.---> | ||
| + | |||
| + | <br clear="all"> | ||
| + | ===Clotho Database Interaction=== | ||
| + | {{Template:ClothoDatabaseFlow}} | ||
| + | |||
| + | <html><font size="6">A</font></html> new concept we want to introduce to the synthetic biology community is this idea of database binding. This computer science influenced concept provides software tools a way to understand biological part databases that can differ across content, ordering and/or architecture. Clotho ''understands'' a standard set of data and its semantic by translating from unstandardized sets of data, and it remembers where unstandardized data came from. This allows Clotho to export unstandardized data to files understood by the community, or to simply take in unstandardized data, manipulate it under a standardized frame, and check in the edited information back to the unstandardized database. The figure to the right illustrates this. This allows Clotho to be many things to many different designers. One can use Clotho as a scheme to import data to new or existing repositories thus allowing the rapid distribution of data, or users may create their own lab environments and use Clotho to help maintain a consistent lab database. | ||
| + | |||
| + | |||
| + | |||
| + | <!---As the synthetic biology community moves toward the standardization of parts and their repositories it is important that a design environment be agnostic when possible to the data it can begin a design with. Clotho allows for the import of both standard and non-standard data. Once the data is brought into the system it can be manipulated, analyzed, modified, and viewed. It then can either be saved back in a non-standard way as it was brought into the system initially or converted to a standardized form and distributed more widely. The figure to the right illustrates such a process.---> | ||
| + | |||
| + | |||
| + | <br clear="all"> | ||
| + | |||
| + | ==Contact Us== | ||
| + | |||
| + | <b>For general information or to contribute:</b> clothodevelopment AT googlegroups DOT com | ||
| + | |||
| + | <b>For emails regarding any bugs:</b> clothobugs AT googlegroups DOT com | ||
| + | |||
| + | <b>For emails regarding any desired features:</b> clothofeatures AT googlegroups DOT com | ||
| + | |||
| + | ==Sponsors== | ||
| + | We thank our Berkeley iGEM Advisory Group: Chris Anderson, Adam Arkin, John Dueber, Jay Keasling, and Susan Marqusee for their support and guidance, our Human Practices Advisors: Gaymon Bennett, Paul Rabinow, and Anthony Stavrianakis, administration support: Kevin Costa and Kate Spohr, and the generous financial support of our sponsors: <br> | ||
| + | [[Image:GSRC.png|200px]] | ||
| + | [[Image:Chess.png|200px]] | ||
| + | [[Image:synberc.jpg|100px]] | ||
| + | [[Image:nsf.jpg|100px]] | ||
| + | <html> | ||
| + | <a href="http://www.gene.com"> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/9/95/Ucb_genentech.jpeg" width="250" height="80"> | ||
| + | </a> | ||
Latest revision as of 22:49, 20 November 2008
With >700 available parts and >2000 defined parts in the MIT Registry of Standard Biological Parts, additional part repositories being developed, and countless more parts local to the lab, synthetic biologists are now faced with the challenge of organizing, sorting, and editing these parts from an ever-growing collection of DNA. Computational tools are becoming increasingly needed to assist biologists to do these jobs, and it is no secret that many tools are being introduced into the community to help serve these needs. It is important that new tool developments in this area remain both firmly rooted in the realities of the present while being forward thinking and flexible. Clotho is a toolset designed in a platform-based design paradigm to consolidate all these tools into one working, integrated toolbox. Clotho is based on a core-and-hub system which manages multiple connections, each connection performing tasks in some self-sufficient manner while connected to all other connections via the core. Examples of connections include sequence viewers/editors, parts database managers, assembly algorithm GUIs, and much more. Computer-savvy biologists can create new connections which will then be integrated into Clotho. These pieces can be kept locally in ones lab or made public and shared with the community. In this tool-consolidation and customization it is hoped that, in the future, biologists need not look further for a program that provides the answer to their ever growing design needs. |
|
Clotho Overview
Among many possible top-level explanations of Clotho, one of the simplest is the "Clotho Data Structure"-centric view. In this view, biological part data is passed back and forth between Clotho components and to files and databases outside Clotho. Some of the possible flows of data are represented by the slew of arrows in the diagram above.
At the Center: Clotho Data Structure
Local and standard databases vary from one lab to another, and the way to grab biological part data from one database may require a completely different route than another. The Clotho Data Structure, in essence, can be viewed as the Rosetta stone of Clotho's environment. In a nutshell, it allows any developer to grab relevant information about a part from any given database. This is made possible when a developer "binds" fields in a database to specific Clotho Data Structure objects which have a specific semantic associated with them. When a user wants to grab the sequence belonging to a particular Biobrick, for example, the Clotho Data Structure, once "bound" to a particular database, will be able to grab the sequence from the database and give it to Clotho. Want to know who owned the vector from jca1021 in database A and send that information to database B? Easy. Bind the data structure with both databases, grab the data from database A, and tell Clotho where in database B to place the data. This binding not only gives unstandardized data meaning, it also provides a common language by which Clotho developers can communicate.
Views/Functions
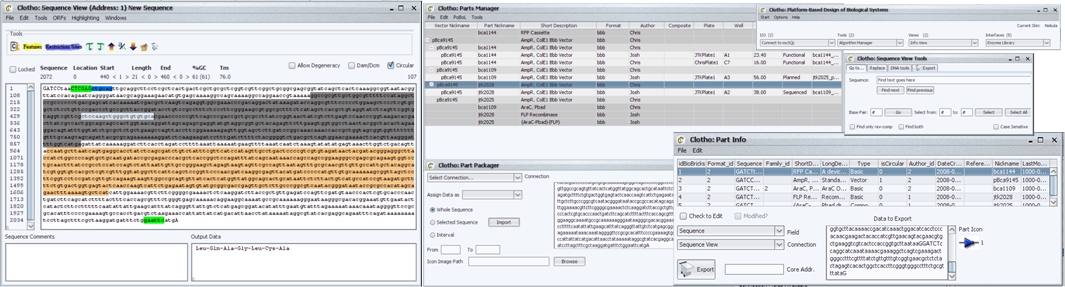
Each category above merits its own explanation, as each is vitally important to the project. Perhaps a good place to start are the views/functions. Here users are able to see and manipulate data, such as part information or the nucleotide sequence. Examples include the sequence view or the plate manager.
Interfaces
Not all views/functions are self-sufficient, and so another set of components, called interfaces, are involved in assisting these views/functions for functionality. Take for example the sequence view (a view many ApE lovers would recognize). How would the sequence view know how to highlight features or enzymes if Clotho didn't know what features or enzymes to look for? That's where the features library and enzyme library come in. In this particular example, the two interfaces store and manage information that the sequence view will have to frequent in order to perform vital operations. Other interfaces include a parts manager and database manager. These allow the user to view part data and relationships. Not only can the source of parts be viewed but users can maintain personal collections of parts as well.
Algorithm Environment
But what if you want to do more with your data you've manipulated? What if, after gathering all your parts, you want to calculate the optimal assembly scheme for a particular goal composite part? This is where the algorithm environment kicks in. The algorithm environment allows developers to make systems that take both quick one-liner inputs or more complex input from the Clotho Data Structure and spit out one-liner or more sophisticated outputs. The algorithm manager allows not only that both approaches can be performed but each are recognized seamlessly. Let's say you wanted to program a partial matching utility into Clotho, where two sequences are given as inputs and a streamlined one-line primer match were spit back out. Algorithm manager provides you all the basic facilities to code such a utility.
Clotho Connectors
Given all these features, there still need to be components that allow Clotho to connect to the outside world. Clotho connectors, so aptly named, perform this exact purpose. These components allow users to connect to databases through internet connections, or import and export data from existing files (think .gb, .ape, .str, etc). Connectors can also translate the results of Clotho back into any number of formats if the user so wishes.
Clotho Plug-in Environment
But last, and certainly not least, is the Clotho plug-in environment. This powerful environment allows developers both associated and not associated with the Clotho Project to create new and customized plug-ins to integrate with their versions of Clotho. Clotho was designed to be as fluid and flexible as possible! While the project is indeed open source, and users can always make new components by making new connections (see Project Technical), developers can now program personalized tools without knowing anything about the Clotho architecture, save for the plug-in environment! Plug-ins are a bit of an intermediate between a Clotho algorithm and a Clotho connection. This allows for relative customization, but also makes the programmer's life easier. They are rooted in the [http://jpf.sourceforge.net/ Java Plugin Framework (JPF)] and should provide a powerful, community driven aspect to Clotho.
We have spent long and hard working hours to develop this tool, and we sure hope you enjoy it. Play around with it and be sure to let us know what you think!
Clotho Database Interaction
A new concept we want to introduce to the synthetic biology community is this idea of database binding. This computer science influenced concept provides software tools a way to understand biological part databases that can differ across content, ordering and/or architecture. Clotho understands a standard set of data and its semantic by translating from unstandardized sets of data, and it remembers where unstandardized data came from. This allows Clotho to export unstandardized data to files understood by the community, or to simply take in unstandardized data, manipulate it under a standardized frame, and check in the edited information back to the unstandardized database. The figure to the right illustrates this. This allows Clotho to be many things to many different designers. One can use Clotho as a scheme to import data to new or existing repositories thus allowing the rapid distribution of data, or users may create their own lab environments and use Clotho to help maintain a consistent lab database.
Contact Us
For general information or to contribute: clothodevelopment AT googlegroups DOT com
For emails regarding any bugs: clothobugs AT googlegroups DOT com
For emails regarding any desired features: clothofeatures AT googlegroups DOT com
Sponsors
We thank our Berkeley iGEM Advisory Group: Chris Anderson, Adam Arkin, John Dueber, Jay Keasling, and Susan Marqusee for their support and guidance, our Human Practices Advisors: Gaymon Bennett, Paul Rabinow, and Anthony Stavrianakis, administration support: Kevin Costa and Kate Spohr, and the generous financial support of our sponsors:





 "
"