Minnesota/8 July 2008
From 2008.igem.org
(Difference between revisions)
Emartin9808 (Talk | contribs) |
|||
| (One intermediate revision not shown) | |||
| Line 8: | Line 8: | ||
{| | {| | ||
|- | |- | ||
| - | |1. '''Simulations:''' Finish typing into | + | |1. '''Simulations:''' Finish typing into the design gui, Hy3S, the simulations/rxn network. NOT USING SYNBIOSS. |
|- | |- | ||
| - | |2. '''Restart/Redo PCR''': Problem with original PCR was an error in the calculation of DNA concentration of product. 07-07-2008 we added only 1.0uL of PCR to digest reactions meaning only 3ng of DNA was in the solution. That was not a substantial amount of DNA to be seen in a electrophoretic gel. FIX - | + | |2. '''Restart/Redo PCR''': Problem with original PCR was an error in the calculation of DNA concentration of product. 07-07-2008 we added only 1.0uL of PCR product to our double digest reactions, meaning only 3ng of DNA was in the solution. That was not a substantial amount of DNA to be seen in a electrophoretic gel, especially considering that only 1/5 of the digest solution was run on the gel and thus only .6 ng of of PCR product DNA was run on our original gel. FIX - Recalculated DNA concentration and concluded that our purified product has a concentration of 3ng/1uL instead of 300ng/uL. At this point, we took two additional steps. We first re-ran the PCR2 product from 7/4, using 13.5 uL of the pure, undigested product. We then redid PCR2 (the addition of the BioBrick prefix to the RBS/Lambda cI gene complex) using 100.0ng of template DNA (the RBS/Lambda cI construct), instead of the 1 ng of DNA which was used on 7/4, by adding 10.0uL of DNA. Once the PCR cycle has completed, we will run the unpurified product on a 1% gel to determine whether or not we have an appropriate product. |
|- | |- | ||
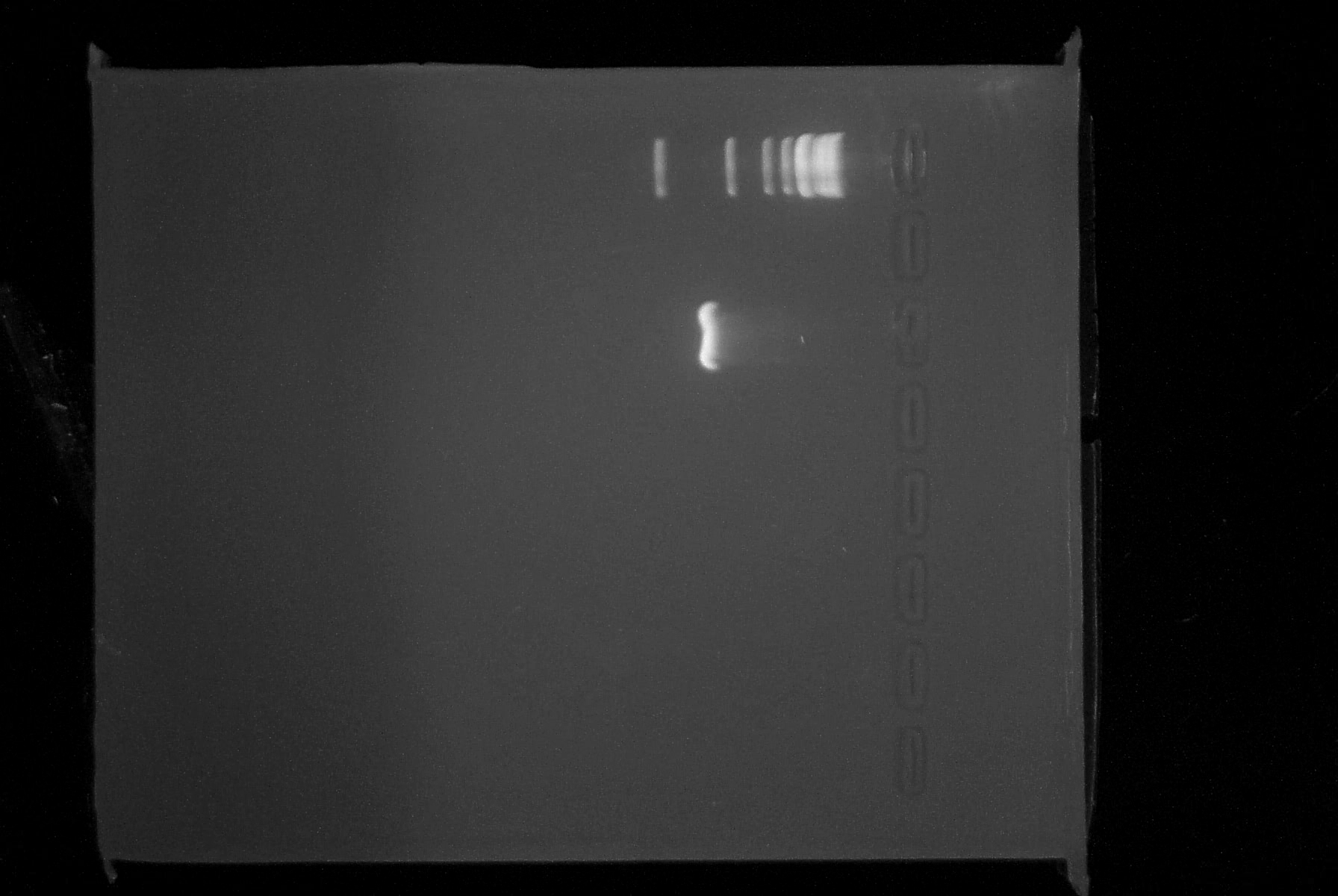
| - | |3. '''PCR''': | + | |3. '''PCR''': It appears that the PCR2 reactions from 7/4 was in fact successful. We saw a faint band at approximately 780 bp, indicating that we had most likely been successful and it was the small amount of DNA which had caused our lack of results yesterday. Additionally, the new PCR reaction was completed successfully. The RBS and BioBrick prefix appear to have been incorporated onto the 5' end of the lambda cI gene. The purified product was spec'ed and has a concentration of 23 ng/uL. Analysis by 1% agarose gel electrophoresis confirmed that the DNA fragment is of the appropriate size (~780 bp). Below is a picture of the gel we ran today. |
| + | |||
| + | Top lane is 1kb ladder, third lane is PCR product. | ||
|- | |- | ||
| - | |[[Image:7.8.08gel.jpg|thumb|left|600px|Gel | + | |[[Image:7.8.08gel.jpg|thumb|left|600px|Gel with ladder and PCR]] |
|} | |} | ||
Latest revision as of 16:00, 9 July 2008
| Back to Notebook Home | |
| Go to Previous Day (July 7) | Go to Next Day (July 9) |
| 1. Simulations: Finish typing into the design gui, Hy3S, the simulations/rxn network. NOT USING SYNBIOSS. |
| 2. Restart/Redo PCR: Problem with original PCR was an error in the calculation of DNA concentration of product. 07-07-2008 we added only 1.0uL of PCR product to our double digest reactions, meaning only 3ng of DNA was in the solution. That was not a substantial amount of DNA to be seen in a electrophoretic gel, especially considering that only 1/5 of the digest solution was run on the gel and thus only .6 ng of of PCR product DNA was run on our original gel. FIX - Recalculated DNA concentration and concluded that our purified product has a concentration of 3ng/1uL instead of 300ng/uL. At this point, we took two additional steps. We first re-ran the PCR2 product from 7/4, using 13.5 uL of the pure, undigested product. We then redid PCR2 (the addition of the BioBrick prefix to the RBS/Lambda cI gene complex) using 100.0ng of template DNA (the RBS/Lambda cI construct), instead of the 1 ng of DNA which was used on 7/4, by adding 10.0uL of DNA. Once the PCR cycle has completed, we will run the unpurified product on a 1% gel to determine whether or not we have an appropriate product. |
| 3. PCR: It appears that the PCR2 reactions from 7/4 was in fact successful. We saw a faint band at approximately 780 bp, indicating that we had most likely been successful and it was the small amount of DNA which had caused our lack of results yesterday. Additionally, the new PCR reaction was completed successfully. The RBS and BioBrick prefix appear to have been incorporated onto the 5' end of the lambda cI gene. The purified product was spec'ed and has a concentration of 23 ng/uL. Analysis by 1% agarose gel electrophoresis confirmed that the DNA fragment is of the appropriate size (~780 bp). Below is a picture of the gel we ran today.
Top lane is 1kb ladder, third lane is PCR product. |
 "
"