Team:EPF-Lausanne/Modeling
From 2008.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
<H3>Simulation</h3> | <H3>Simulation</h3> | ||
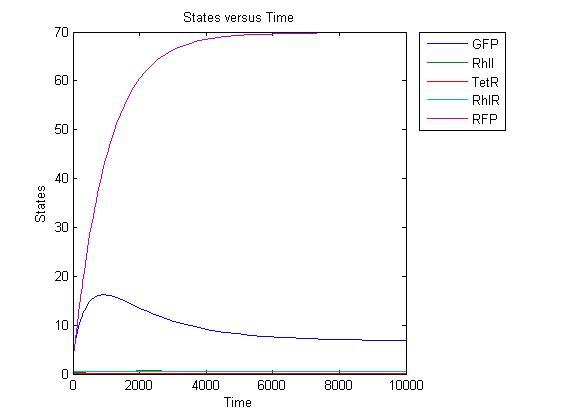
| - | <p> Simulation has began. We have | + | <p> Simulation has began. We have built a model using SimBiology. The ODEs have all been computed. The main problem now resides in definining the parameters correctly. |
| - | Primary results look | + | Primary results look promising. An RhlI-induced reaction gave the desired curves, well nearly. GFP still ends up too high. The other main problem is going to be the response time. But we have to wait until we have a complete and robust model to really start finding solutions to our problems. |
</p> | </p> | ||
<table border="0"> | <table border="0"> | ||
| Line 10: | Line 10: | ||
<td> | <td> | ||
<p> | <p> | ||
| - | The simulation starts | + | The simulation starts off with constitutive expression of GFP. After RhlI "injection", GFP production slows down and RFP starts to be expressed. Speeds and concentrations are not yet optimal but this was a quick first try to check if the model was viable. Conclusion: OK. |
</p> | </p> | ||
</td> | </td> | ||
Revision as of 13:26, 28 July 2008
Simulation
Simulation has began. We have built a model using SimBiology. The ODEs have all been computed. The main problem now resides in definining the parameters correctly. Primary results look promising. An RhlI-induced reaction gave the desired curves, well nearly. GFP still ends up too high. The other main problem is going to be the response time. But we have to wait until we have a complete and robust model to really start finding solutions to our problems.
Have been considering LacI and LacIm as two different proteins. But after amino acid comparaison they are actually the same. Only the promoter and DNA sequenses are different.
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Note
The Ordinary Differential Equations (ODE) of our system are listed in the following file :
 "
"