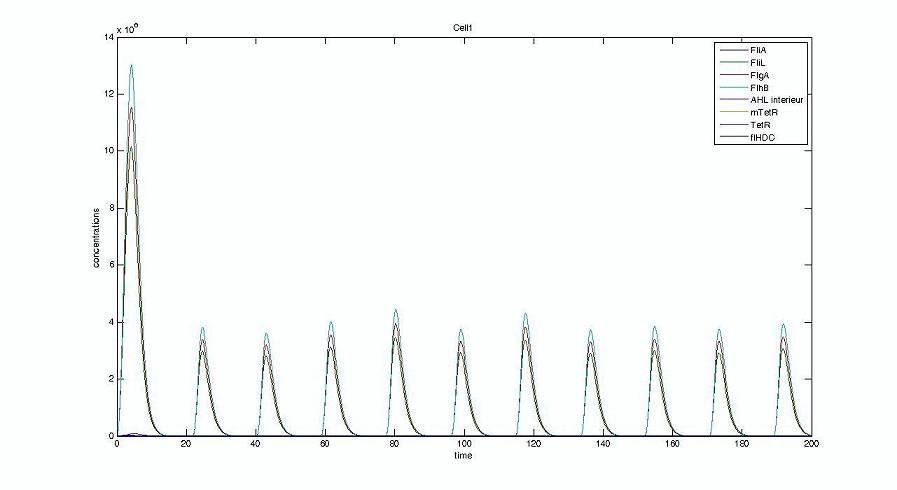
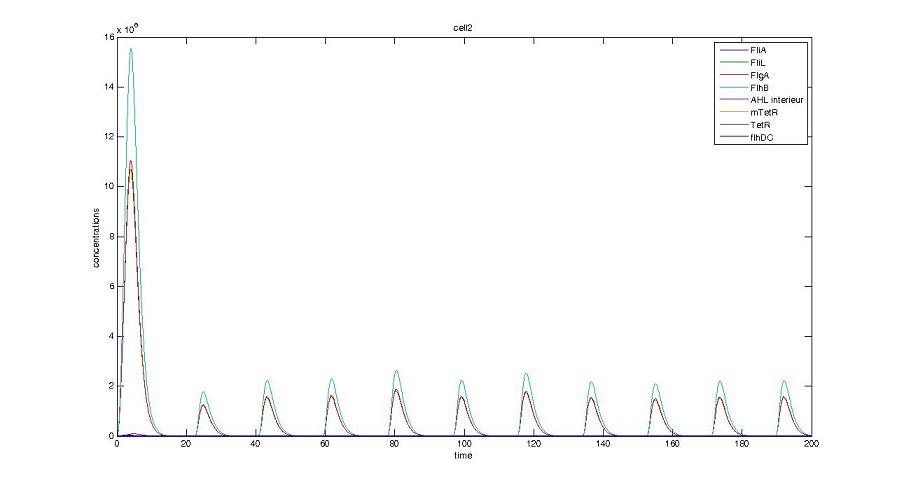
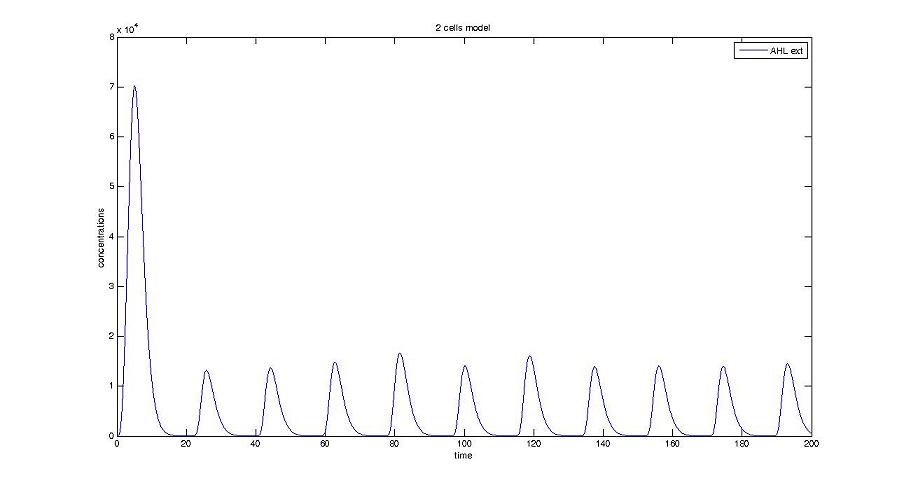
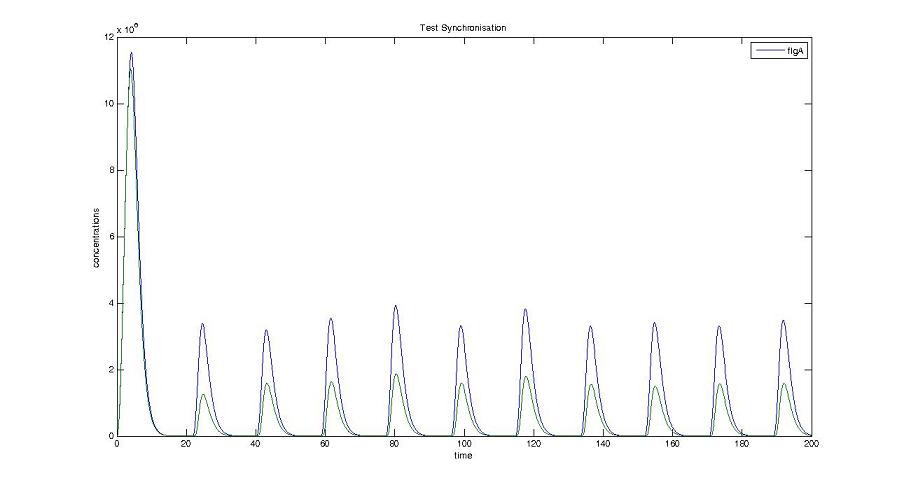
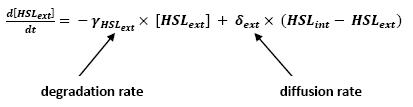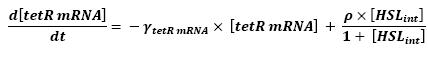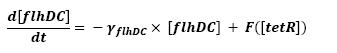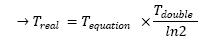Too be continued
From 2008.igem.org
(→Equations) |
(→First Mathematical Approach) |
||
| (6 intermediate revisions not shown) | |||
| Line 111: | Line 111: | ||
<br> | <br> | ||
| - | </br> <font color=blue>Again, it is not at all a trivial fact that the equations here are linear. Discuss.</ | + | </br> <font color=blue>Again, it is not at all a trivial fact that the equations here are linear. Discuss.</font> |
---- | ---- | ||
* <b>Steps</b> | * <b>Steps</b> | ||
| Line 121: | Line 121: | ||
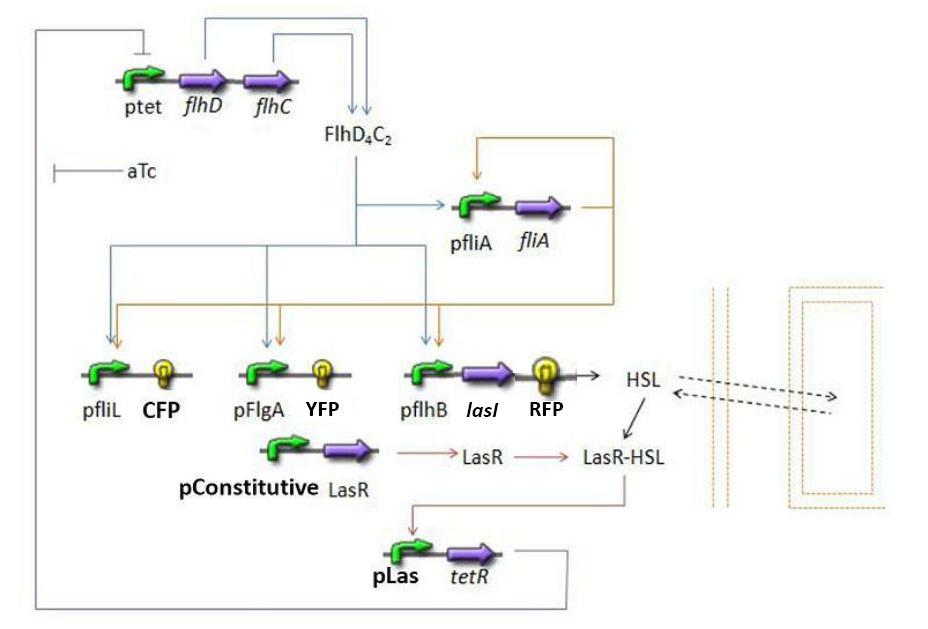
[[Image:HSLext.jpg|center]] | [[Image:HSLext.jpg|center]] | ||
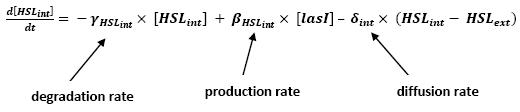
| - | * <b> | + | * <b>Discussion and Assumptions</b> |
** In a first approach, we have assumed that HSL could be modelized in the same fashion as AHL. The process was well detailed in [[Team:Paris/Modeling#Bibliography|[2]]] and we adapted their model to our present situation. Furthermore, a biologically coherent set of parameters is also given in this article. We decided then to use them as well for our simulation. | ** In a first approach, we have assumed that HSL could be modelized in the same fashion as AHL. The process was well detailed in [[Team:Paris/Modeling#Bibliography|[2]]] and we adapted their model to our present situation. Furthermore, a biologically coherent set of parameters is also given in this article. We decided then to use them as well for our simulation. | ||
| + | <br> <font color=blue> It is good to give the reference, but for the readability of your presentation you should include here also briefly the justification of your model. Too rapid just to refer to the bibliography.</font> | ||
<br> | <br> | ||
| Line 135: | Line 136: | ||
[[Image:tetR.jpg|center]] | [[Image:tetR.jpg|center]] | ||
| - | * <b> | + | * <b>Discussion and Assumptions</b> |
** Here is the only case where we have not yet found out how we could manage to skip the mRNA step. However, this is acceptable since every parameter should be available. Then, (12) represents the binding and transcription steps ; (13) represents the translation step. | ** Here is the only case where we have not yet found out how we could manage to skip the mRNA step. However, this is acceptable since every parameter should be available. Then, (12) represents the binding and transcription steps ; (13) represents the translation step. | ||
| + | |||
| + | <br> <font color=blue> Could you discuss why? It is not clear to me straight on.</font> | ||
** The effect of HSL as well as the link between the mRNA activity and protein activity is well studied in [[Team:Paris/Modeling#Bibliography|[2]]]. We were the able to adapt the introduced situation to our system. | ** The effect of HSL as well as the link between the mRNA activity and protein activity is well studied in [[Team:Paris/Modeling#Bibliography|[2]]]. We were the able to adapt the introduced situation to our system. | ||
** The ρ parameter corresponds to maximal contribution to tetR transcription in presence of saturating amounts of HSL. Yet, to begin with, we have used the parameter given in the text. We shall then find a more accurate parameter corresponding to our system. The HSL activation is hereby chosen to follow Michalelis Menten kinetics since we assumed that there would be no cooperativity. | ** The ρ parameter corresponds to maximal contribution to tetR transcription in presence of saturating amounts of HSL. Yet, to begin with, we have used the parameter given in the text. We shall then find a more accurate parameter corresponding to our system. The HSL activation is hereby chosen to follow Michalelis Menten kinetics since we assumed that there would be no cooperativity. | ||
| Line 150: | Line 153: | ||
[[Image:flhDC.jpg|center]] | [[Image:flhDC.jpg|center]] | ||
| - | * <b> | + | * <b>Discussion and Assumptions</b> |
** To determine this part of the model, we used [[Team:Paris/Modeling#Bibliography|[4]]] where the influence of tetR is described. | ** To determine this part of the model, we used [[Team:Paris/Modeling#Bibliography|[4]]] where the influence of tetR is described. | ||
** Finally, to close the loop, we saw the necessity to use a little refinment. the influence of tetR over pTet (pTet being the promoter of flhDC) is a repression. To express the fact that a certain concentration is needed for the expression of flhDC, we used a step function. This choice was influenced by [[Team:Paris/Modeling#Bibliography|[3]]]. To fully define this function, two parameters are needed, so as to describe the threshold when the step has to occur, and the initial value of this step. We are facing two solutions. The more accurate procedure would consist in getting those values from the wet-lab. However, we may also look it up in the literature. Anyhow, as presented in the conclusive part, there is a really wide range of parameters that enable the system to oscillate. | ** Finally, to close the loop, we saw the necessity to use a little refinment. the influence of tetR over pTet (pTet being the promoter of flhDC) is a repression. To express the fact that a certain concentration is needed for the expression of flhDC, we used a step function. This choice was influenced by [[Team:Paris/Modeling#Bibliography|[3]]]. To fully define this function, two parameters are needed, so as to describe the threshold when the step has to occur, and the initial value of this step. We are facing two solutions. The more accurate procedure would consist in getting those values from the wet-lab. However, we may also look it up in the literature. Anyhow, as presented in the conclusive part, there is a really wide range of parameters that enable the system to oscillate. | ||
| + | |||
| + | <br> <font color=blue>What is the function you are using for F()? </font> | ||
** The key problem was that those two parameters were to be determined by the overall evolution of tetR, and the program cannot wait for tetR to be entirely determined to evaluate flhDC, since we have a loop. We then decided that these parameters could be obtanied by lab experiments. | ** The key problem was that those two parameters were to be determined by the overall evolution of tetR, and the program cannot wait for tetR to be entirely determined to evaluate flhDC, since we have a loop. We then decided that these parameters could be obtanied by lab experiments. | ||
| Line 435: | Line 440: | ||
* In [[Team:Paris/Modeling#Bibliography|[1]]], we had trouble deciding whether the parameters we used were coherent or not, and we spent some time checking abouts units. Thus, we had to decide how we shall set our time unit. | * In [[Team:Paris/Modeling#Bibliography|[1]]], we had trouble deciding whether the parameters we used were coherent or not, and we spent some time checking abouts units. Thus, we had to decide how we shall set our time unit. | ||
* We found a seemingly acceptable answer using elements from [[Team:Paris/Modeling#Bibliography|[1]]] and [[Team:Paris/Modeling#Bibliography|[5]]]. | * We found a seemingly acceptable answer using elements from [[Team:Paris/Modeling#Bibliography|[1]]] and [[Team:Paris/Modeling#Bibliography|[5]]]. | ||
| + | |||
| + | |||
| + | <br> <font color=blue>Good section for discussing relationship between model quantities and whole population measurements. You should explain better the purpose of this part however. What are the conditions of measurement. Why the optical density. What is $GFP_{1cell}$. Protein level activity is the concentrationof the protein? What is $[Y]$. Describe more how to get the final relationship between $T_{equation}$ and $T_{real}$.</font> | ||
| + | |||
[[Image:raisonnement math.jpg|center]] | [[Image:raisonnement math.jpg|center]] | ||
| Line 461: | Line 470: | ||
* This enabled us to set a real x-axis unit (we had decided for practical reasons to set α to 1). | * This enabled us to set a real x-axis unit (we had decided for practical reasons to set α to 1). | ||
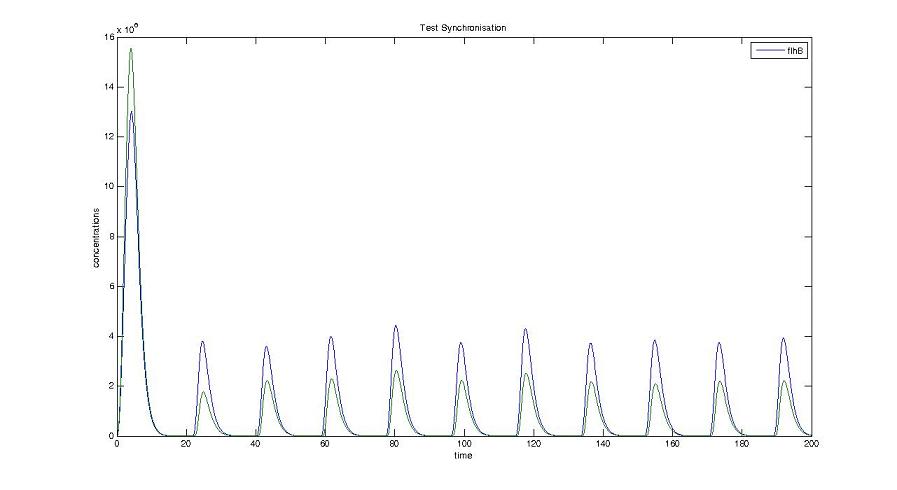
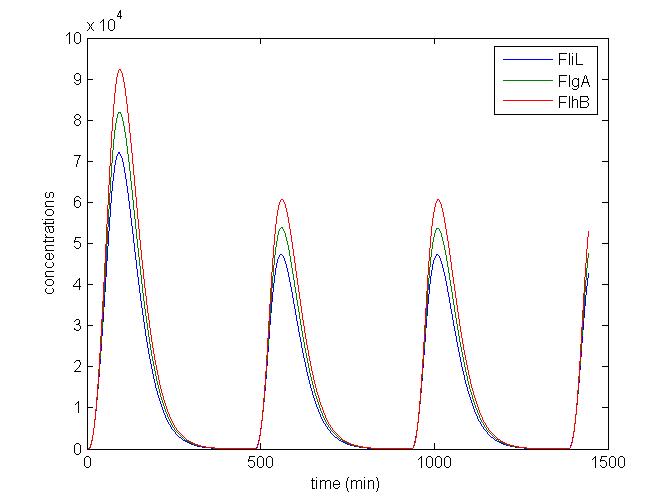
* Finally, we have compared our results with the experiments carried out in [[Team:Paris/Modeling#Bibliography|[5]]]. It appears that the timescales to turn the genes 'ON' are similar ! | * Finally, we have compared our results with the experiments carried out in [[Team:Paris/Modeling#Bibliography|[5]]]. It appears that the timescales to turn the genes 'ON' are similar ! | ||
| + | |||
| + | <br> <font color=blue>Explain the comparison. It is not clear where are the time scales you compared.</font> | ||
[[Image:Real_time_oscill.jpg|center]] | [[Image:Real_time_oscill.jpg|center]] | ||
| Line 474: | Line 485: | ||
===Introduction=== | ===Introduction=== | ||
| + | |||
| + | <br> <font color=blue>This description of the approaches should come earlier. It is not clear reading you if you are speaking of the model presented above, or of something else you did before. I think this part that presents your work on modeling should come before the detailed developments of above. In the introduction to help the reader understand where you are heading. </font> | ||
As a first approach, we had decided to take into account the binding to the promoters steps. Moreover, the transcription rates were expected to be Hill functions. Obvisouly, this modeling requires a huge number of parameters. To obtain them, we had planed to devise specific experiments (described below). | As a first approach, we had decided to take into account the binding to the promoters steps. Moreover, the transcription rates were expected to be Hill functions. Obvisouly, this modeling requires a huge number of parameters. To obtain them, we had planed to devise specific experiments (described below). | ||
| - | Nonetheless, after reading some more articles, we have decided to change several | + | Nonetheless, after reading some more articles, we have decided to change several assumptions of the modeling choice. Therefore, we have devised a perhaps more biologically relevant framework (see above). |
This part describes in detail the first approach and the codes that have been produced. | This part describes in detail the first approach and the codes that have been produced. | ||
Latest revision as of 12:35, 12 August 2008
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"