Team:Caltech/Protocols/PABA HPLC assay
From 2008.igem.org
(→Standard Curve) |
(→Standard Curve) |
||
| Line 45: | Line 45: | ||
===Standard Curve=== | ===Standard Curve=== | ||
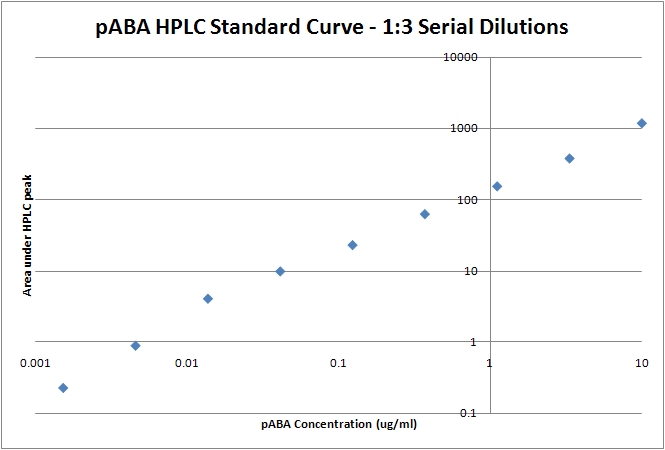
| - | 1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol. | + | 1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol to generate a linear standard (see below). |
| - | [[Image:08-07-08_pABA_Standard_Curve_plot.jpg | pABA HPLC standard curve with 1:3 dilutions starting from 10ug/ml pABA. The integral of the peak is plotted against the known pABA amount.]] | + | [[Image:08-07-08_pABA_Standard_Curve_plot.jpg | thumb|left |pABA HPLC standard curve with 1:3 dilutions starting from 10ug/ml pABA. The integral of the peak is plotted against the known pABA amount.]] |
==Sources== | ==Sources== | ||
Guo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. ''Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry.'' '''Rapid Communications in Mass Spectrometry''': Volume 19, Issue 8 , Pages 963 - 969, 2005. | Guo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. ''Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry.'' '''Rapid Communications in Mass Spectrometry''': Volume 19, Issue 8 , Pages 963 - 969, 2005. | ||
Revision as of 08:30, 23 August 2008
|
People
|
{{{Content}}} |
</div>
para-Aminobenzoic Acid (pABA) HPLC Assay ProtocolPurposeTo test for para-aminobenzoic acid levels in E. coli overexpressing the pABA synthesis genes pabA and pabB. Materials
ProcedureSample Prep1. Centrifuge the cell culture max speed, 10 minutes. 2. Separate pellet from supernatant. 3. Resuspend the pellet in 1mL 0.1M Tris-HCl buffer 4. Sonicate the resuspended pellet for 1 minute, alternating between 0 and 12Hz. 5. Filter sterilize the cell lysate and the supernatant. 6. Load 500uL of each into the HPLC. HPLC Method1. Injection volume was 20uL, column temperature was kept at 40°C. 2. Flush the lines individually with Buffer A and B for two minutes each, flush the column with 92% A, 8% B for 10 minutes. 3. From the source: "The starting eluent was 92% A mixed with 8% B; the proportion of B was increased linearly to 50% in 7 min, then to 100% in 3 min. The mobile phase was then immediately adjusted to its initial composition and held for 4 min in order to re-equilibrate the column." 4. We found the retention time of pABA to be 4.9 minutes, however the literature states the retention time as 6.0 minutes. Standard Curve1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol to generate a linear standard (see below). SourcesGuo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry. Rapid Communications in Mass Spectrometry: Volume 19, Issue 8 , Pages 963 - 969, 2005. |
 "
"