Imperial College/2 September 2008
From 2008.igem.org
(Difference between revisions)
m (→Results) |
m (→Testing of Primers) |
||
| Line 32: | Line 32: | ||
**15-47 <sup>o</sup>C - Negative control - no DNA!!! | **15-47 <sup>o</sup>C - Negative control - no DNA!!! | ||
**16-51 <sup>o</sup>C - AmyE 1 positive control as previously shown to work. | **16-51 <sup>o</sup>C - AmyE 1 positive control as previously shown to work. | ||
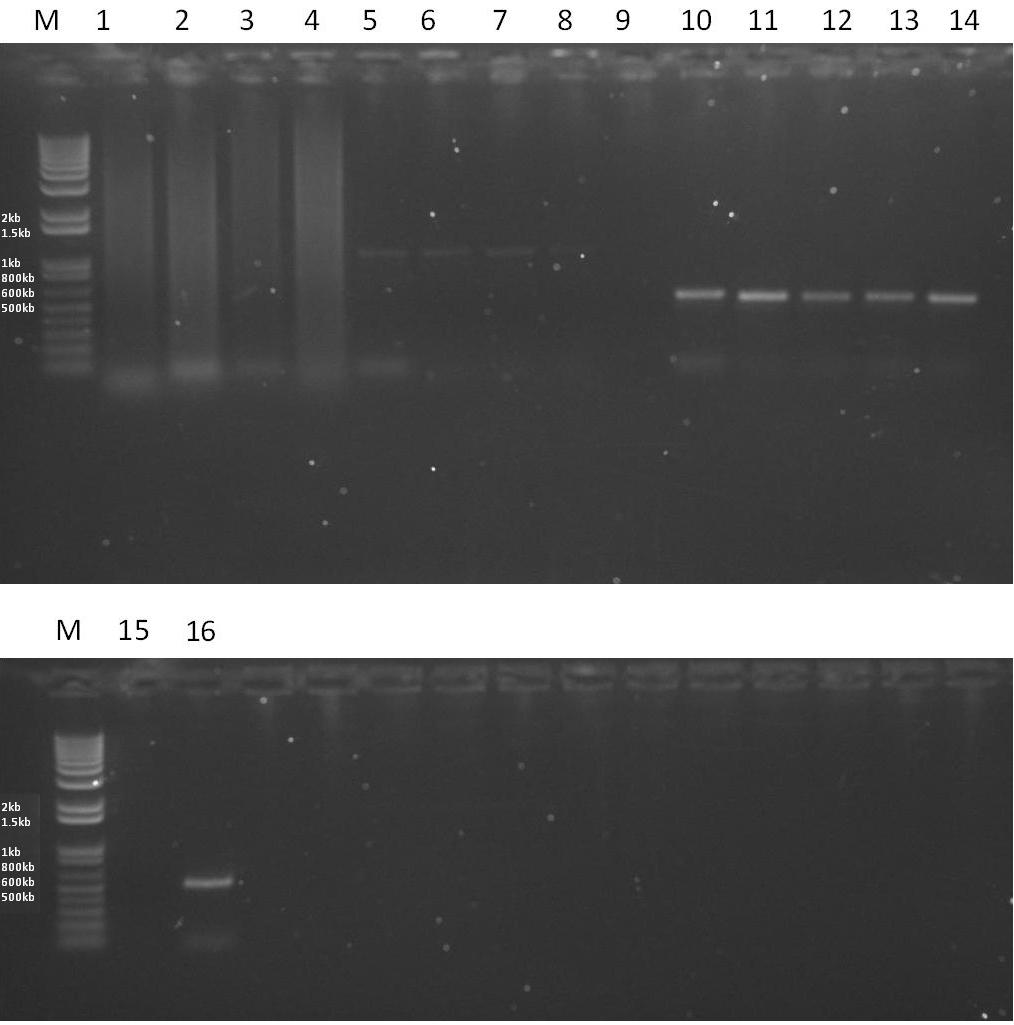
| - | + | *'''Results''' - From the gel below it can be seen that the EpsE PCR reactions have worked, however with slight levels of contamination when a high annealing temperature is used. The XylR PCR reaction has not work efficiently, clearly visible from the faint bands. This means that the temperatures for annealing need to be optimised further. | |
===Results=== | ===Results=== | ||
[[Image:RESULTS2.JPG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer.]] | [[Image:RESULTS2.JPG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer.]] | ||
Revision as of 17:56, 2 September 2008
Wet LabTransformation of B.subtilis
Testing of Primers
Results |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"