Team:Caltech/Project/Oxidative Burst
From 2008.igem.org
| Line 7: | Line 7: | ||
__NOTOC__ | __NOTOC__ | ||
===The General Idea=== | ===The General Idea=== | ||
| - | In order to help guard against infections of the gut, we wish to engineer a strain of E. coli capable of killing bacterial pathogens. White blood cells (neutrophils in particular) are already very efficient at killing bacteria. | + | In order to help guard against infections of the gut, we wish to engineer a strain of E. coli capable of killing bacterial pathogens. White blood cells (neutrophils in particular) are already very efficient at killing bacteria. Neutrophils kill bacteria by exposing it to a bombardment of reactive oxygen species including superoxide, hydrogen peroxide and hydrochlorous acid. The reactive oxygen species kill the bacteria by shredding biological molecules they come in contact with by way of their potent oxidizing properties. However neutrophils are not able to migrate to the large intestinal lumen where pathogens can reside. Because bacteria are well adapted to live in the gut, this project’s goal is to engineer a strain of E. coli to detect and kill invading bacterial pathogens by means of a sudden burst of hydrogen peroxide. |
===Detection=== | ===Detection=== | ||
Revision as of 19:51, 16 October 2008
|
People
|
Pathogen Defense by Oxidative Burst
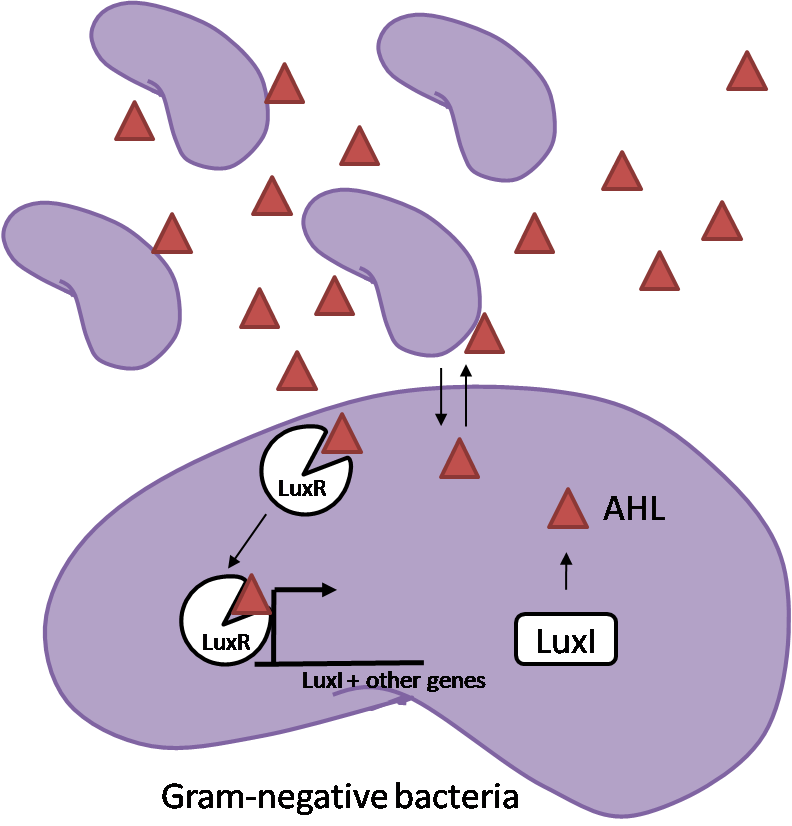
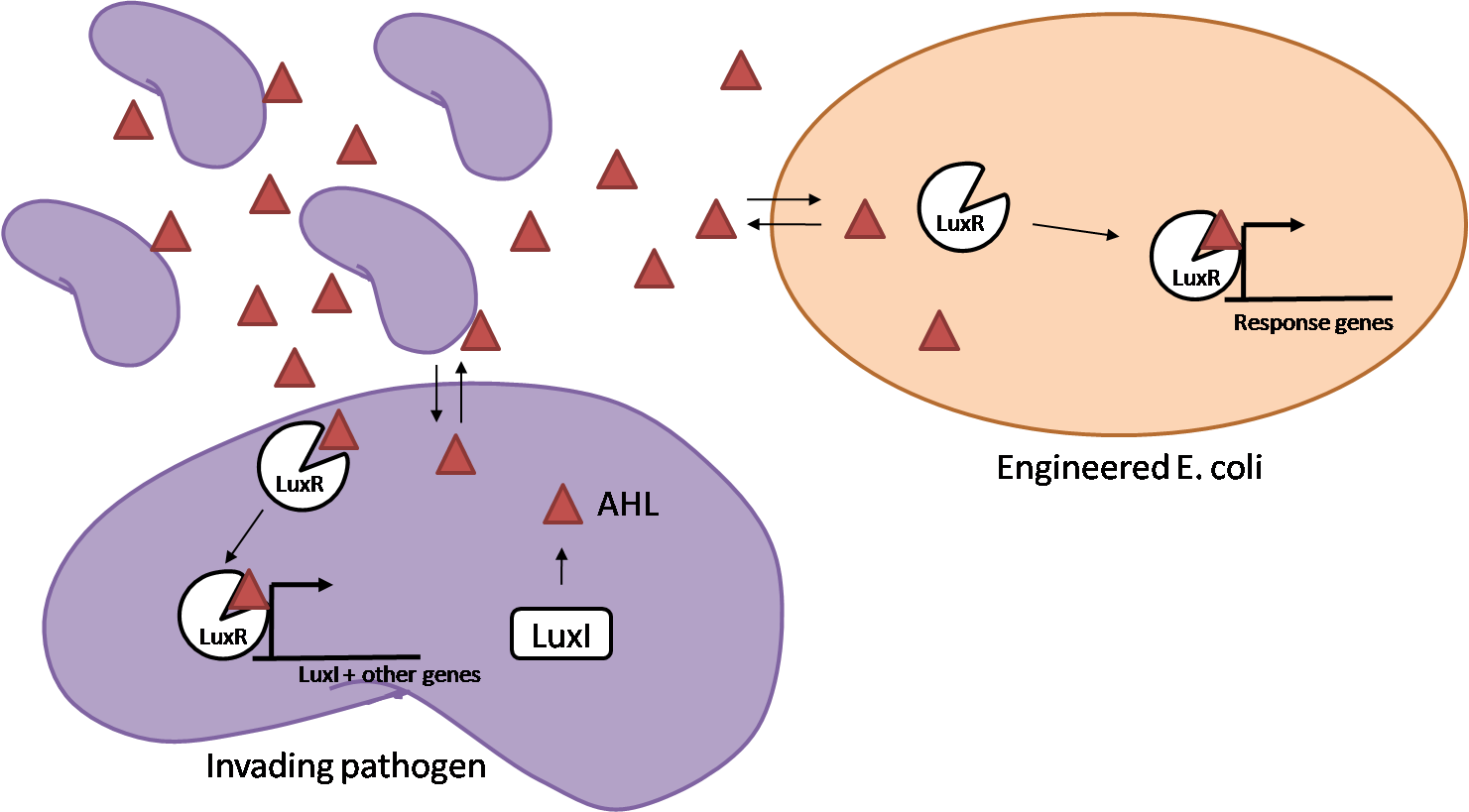
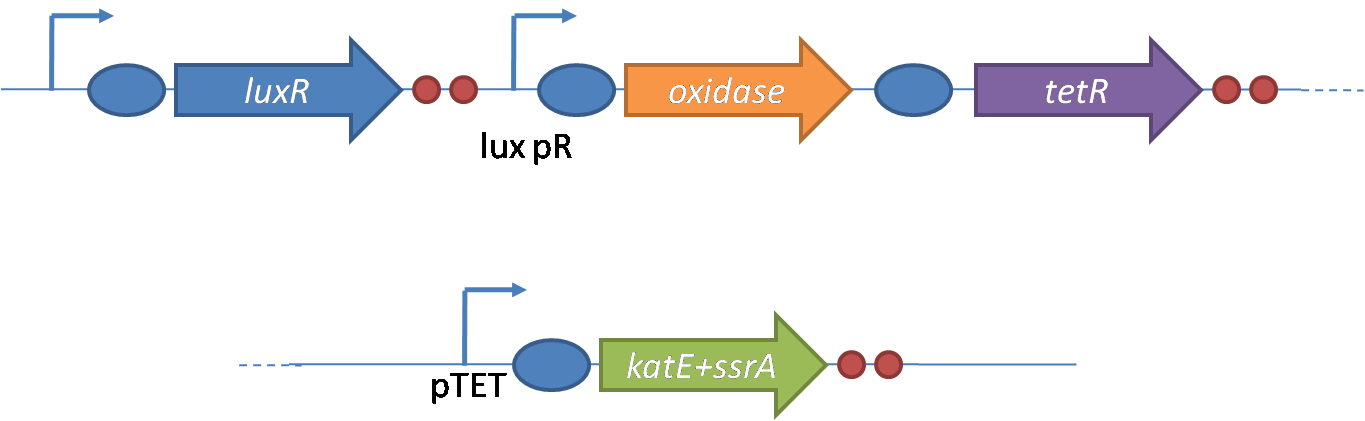
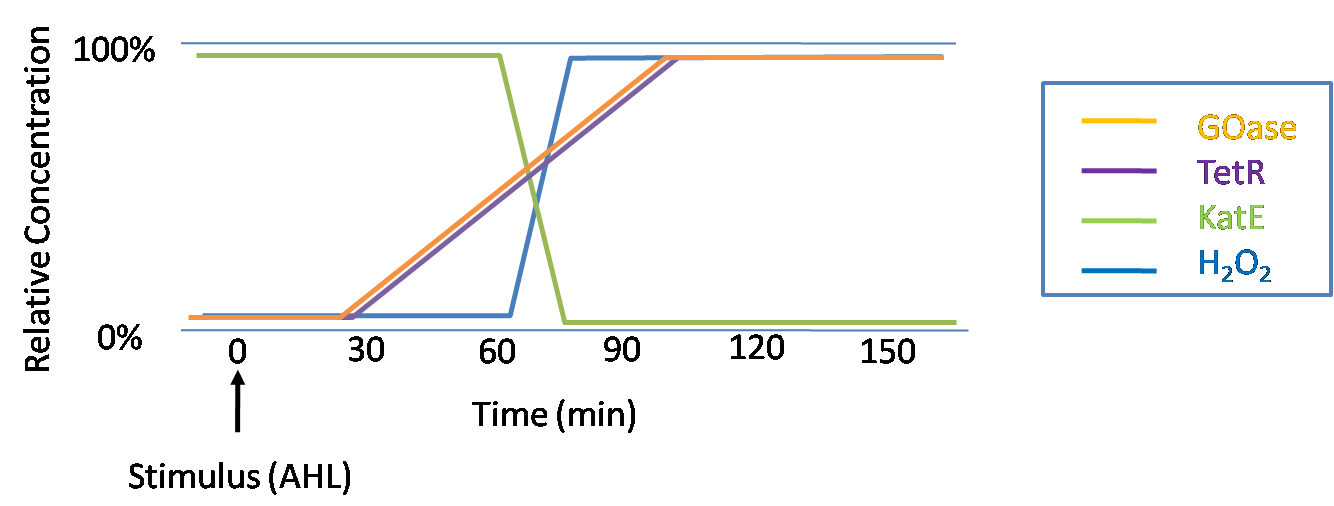
The General IdeaIn order to help guard against infections of the gut, we wish to engineer a strain of E. coli capable of killing bacterial pathogens. White blood cells (neutrophils in particular) are already very efficient at killing bacteria. Neutrophils kill bacteria by exposing it to a bombardment of reactive oxygen species including superoxide, hydrogen peroxide and hydrochlorous acid. The reactive oxygen species kill the bacteria by shredding biological molecules they come in contact with by way of their potent oxidizing properties. However neutrophils are not able to migrate to the large intestinal lumen where pathogens can reside. Because bacteria are well adapted to live in the gut, this project’s goal is to engineer a strain of E. coli to detect and kill invading bacterial pathogens by means of a sudden burst of hydrogen peroxide. DetectionBacteria are able to communicate between individuals of the same species by way of quorum sensing. Small molecules serve as the signal between individual cells. Gram negative bacteria use acylhomoserine lactones (AHL), which can freely diffuse across the cell membrane. The quorum sensing machinery relies in two enzymes, LuxI, an AHL producer, and LuxR, an AHL-dependent transcriptional activator. Figure 1 illustrates how the system works. In isolation, each bacterium constitutively produces a small amount of AHL, which quickly diffuses into the surrounds. If other bacteria of the same species are also nearby, the AHL will diffuse across their membrane where it will bind to LuxR. LuxR activates transcription of several genes, including luxI. A positive feedback loop is created, in which more AHL induces more LuxI, which in turn produces more AHL. Each species of gram negative bacteria produces a unique AHL, requiring unique LuxI and LuxR proteins, and so avoids crosstalk between species. A group of bacteria can thus toggle between an “off” state and an “on” state by using quorum sensing. Our engineered strain will not participate directly in quorum sensing, but instead will eavesdrop on the conversation. It will be engineered to constitutively express a LuxR able to detect a species AHL. In this way, our engineered strain will be able to be tuned to specifically respond to a variety of bacterial pathogens. Once the AHL is bound, LuxR will activate a set of genes which will lead to the overproduction of hydrogen peroxide, killing the invading cell. ResponseAfter sensing the presence of an invading pathogen, we want to engineer our E. coli to produce lethal amounts of hydrogen peroxide relatively quickly. We are not concerned with having the engineered E. coli survive either, as it is reasonable to assume that there are “unactivated” cells far away from the pathogen that could sustain the population. After LuxR binds AHL, it will activate transcription of an oxidase (an enzyme that produces hydrogen peroxide). There are many enzymes that can produce hydrogen peroxide in a stoichiometric ratio. The enzyme currently being used is galactose oxidase, as it has already undergone directed evolution to optimize its thermo stability and has been shown to be highly active in vitro. Other enzymes being considered are pyruvate oxidase and aldehyde oxidase. Even though our engineered cells will eventually die from their oxidative burst, we want them to survive long enough to produce large amounts of the oxidase so they can produce large amounts of hydrogen peroxide. If the cells were left to produce hydrogen peroxide without any protection, they would produce just enough to be cytotoxic and then fissle, killing only themselves but not much else. To avoid this problem, an E. coli catalase will be constitutively expressed, and then turned off shortly after the oxidase is being expreseed. We’re accomplishing this by putting katE (on of two E. coli catalase genes) behind the tetR sensitive promoter (tetR P) and having tetR co-transciptionally expressed with the oxidase. The time it takes for tetR to accumulate in the cell provides the delay in repressing katE expression. To ensure katE is rapidly cleared from the cells, it has a C-terminal ssrA degradation tag, which should reduce the protein’s half life to the order of minutes. In this way, the cell can be temporarily protected from hydrogen peroxide, but large amounts can accumulate before the substrate is exhausted.The final strain will have deletions of both catalases, ensuring no interference. System DesignThe entire system will be engineered onto a high copy plasmid in E. coli as seen in Figure 3. Promoters are shown as bent, thin arrows, genes as thick arrows, ribosome binding sites as blue ovals, and a double transcriptional terminator as two red circles. Initially, the system will function independently, with luxR under a constitutive promoter. Later, luxR will be put under the control of a recombinase system that will only turn on the pathway randomly in a subset of cells. This will be one of three fates the master strain will be able to differenciate into. For more information, visit the population variation page. An ideal system would behave as outlined in Figure 4. It shows the relative abundances of the proteins and molecules used in the system. It is only meant to show if a particular species is relatively high or low, not its absolute concentration. Current Progress
References |
 "
"