Team:Caltech/Project/Phage Pathogen Defense
From 2008.igem.org
| Line 7: | Line 7: | ||
__NOTOC__ | __NOTOC__ | ||
==Introduction== | ==Introduction== | ||
| - | [[Image:PathogenicEcoli.jpg|thumb|left|Pathogenic E. coli.]] | + | [[Image:PathogenicEcoli.jpg|thumb|left|Pathogenic ''E. coli''.]] |
There are <math> 10^{14} </math> bacterial cells that naturally reside in the human gut, an order of magnitude greater than all the cells in the body. Humans enjoy a mutualistic relationship with intestinal microbiota, wherein the microorganisms perform a host of useful functions, such as processing unused energy substrates, training the immune system, and inhibiting growth of harmful bacterial species [1]. However, not all bacteria populations within the human intestine are benign. There are many pathogenic bacteria which cause a wide variety of diseases when present in the human gut. As one example, cholera is one of the most widespread and damaging of such infectious microorganisms, with the World Health Organization reporting 132,000 cases in 2006 [2]. But cholera is just one of many examples. In the United States alone, there were more than 325,000 hospitalizations from food-borne illnesses in the year of 2006 [3] | There are <math> 10^{14} </math> bacterial cells that naturally reside in the human gut, an order of magnitude greater than all the cells in the body. Humans enjoy a mutualistic relationship with intestinal microbiota, wherein the microorganisms perform a host of useful functions, such as processing unused energy substrates, training the immune system, and inhibiting growth of harmful bacterial species [1]. However, not all bacteria populations within the human intestine are benign. There are many pathogenic bacteria which cause a wide variety of diseases when present in the human gut. As one example, cholera is one of the most widespread and damaging of such infectious microorganisms, with the World Health Organization reporting 132,000 cases in 2006 [2]. But cholera is just one of many examples. In the United States alone, there were more than 325,000 hospitalizations from food-borne illnesses in the year of 2006 [3] | ||
There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this fact, dietary supplements have been popularized which aim to introduce beneficial bacteria back into the gut after antibiotic treatment; these substances have been termed ‘probiotics’. Natural probiotics have two main advantages: they provide useful functions for the host and they competitively inhibit growth of pathogenic bacteria. | There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this fact, dietary supplements have been popularized which aim to introduce beneficial bacteria back into the gut after antibiotic treatment; these substances have been termed ‘probiotics’. Natural probiotics have two main advantages: they provide useful functions for the host and they competitively inhibit growth of pathogenic bacteria. | ||
| - | Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to create a novel probiotic with improved medical applications, this project focuses on engineering a pathogen defense system within E. coli.. | + | Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to create a novel probiotic with improved medical applications, this project focuses on engineering a pathogen defense system within ''E. coli''.. |
| + | ===System Design=== | ||
| + | The engineered probiotic acts as a delivery vehicle for phage into the large intestine. There are four important design goals for such as system: (1) the system releases phage into the large intestine, (2) the production of phage is regulated with a high dynamic range between a distinct on and off state, (3) the system can target a wide range of pathogenic bacteria, and (4) the system integrates well within the existing iGEM probiotic. | ||
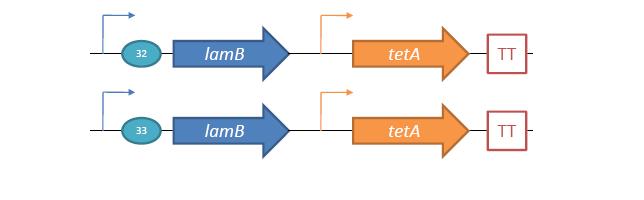
| - | + | An obvious delivery mechanism would be the adaptation of natural lysogens, a phase of the phage life cycle in which the phage integrates its DNA with the host organism. Lysogens lie dormant until disturbed by some external stimuli, but can be stimulated to enter the lytic cycle and release phage. There is one major limitation to this method: phage released will be specific to the lysogenic hosts, ''E. coli''. We can avoid this limitation by incorporating as a lysogen a phage which is not normally infectious to ''E. coli''. This phage could potentially target pathogenic bacteria besides ''E. coli''. To demonstrate the feasibility of this approach, a simple model system was developed using λ phage and JW3996-1, a strain of ''E. coli'' immune to λ infection due to ''lamB'' deletion[5]. In this model, the host is immune to infection unless lamB is expressed on a plasmid. After infection and selection for lysogens, the ''lamB'' plasmid is counterselected against to regain immunity. Extending this model to a more realistic situation, we envision using a phage which targets a pathogenic bacterial species, constitutively expressing the phage receptor within ''E. coli'' to create lysogens, and inducing the lysogens to infect non-''E. coli'' pathogenic bacteria. | |
| - | + | ||
| - | |||
| - | |||
[[Image:SystemDesign 1.jpg |frame|center|50px|Design for the lamB+tetA constructs used for lambda infection and tetracycline counterselection.]] | [[Image:SystemDesign 1.jpg |frame|center|50px|Design for the lamB+tetA constructs used for lambda infection and tetracycline counterselection.]] | ||
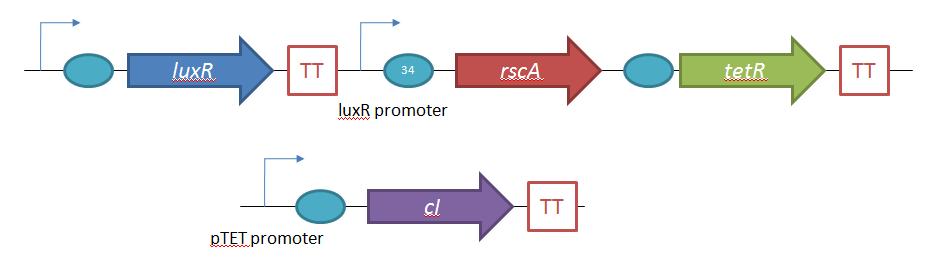
| - | The second plasmid which must be designed controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. | + | The second plasmid which must be designed controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. The induction of λ phage lysogens has been explored since 1950 [6], and serves as a model system for the study of other temperate phages. The most famous induction mechanism is ultraviolet radiation (UV). In UV induction of λ lysogens, UV induced DNA damage activates the cellular SOS mechanism, which activates the expression of RecA, a protein involved in SOS response. RecA, in turn, cleaves the λ repressor and leads to phage activation. In this study, activation of the ''recA'' pathway was not readily feasible, so an alternative effector was used to elicit induction. A secondary plasmid expressing the ''E. coli'' regulatory gene, ''rcsA'', is used as the trigger for λ phage induction. ''rcsA'' is one of several regulatory genes which provide a ''recA'' independent pathway for λ induction[7]. Overexpression of ''rcsA'' leads to λ induction, and, furthermore, ''rcsA'' appears to directly compete with the λ suppression gene ''cI''. [6] In this project, ''rcsA'' is used to trigger phage production. The effectiveness and dynamic range of ''rcsA'' induction is explored. |
[[Image:SystemDesign 2.jpg |thumb|center|650px|Design for the control system for control of lambda phage lysogenic/lytic life cycles.]] | [[Image:SystemDesign 2.jpg |thumb|center|650px|Design for the control system for control of lambda phage lysogenic/lytic life cycles.]] | ||
| + | |||
==Part II: B. Subtilis Lysogens== | ==Part II: B. Subtilis Lysogens== | ||
Revision as of 05:07, 23 October 2008
|
People
|
Phage Pathogen Defense
IntroductionThere are <math> 10^{14} </math> bacterial cells that naturally reside in the human gut, an order of magnitude greater than all the cells in the body. Humans enjoy a mutualistic relationship with intestinal microbiota, wherein the microorganisms perform a host of useful functions, such as processing unused energy substrates, training the immune system, and inhibiting growth of harmful bacterial species [1]. However, not all bacteria populations within the human intestine are benign. There are many pathogenic bacteria which cause a wide variety of diseases when present in the human gut. As one example, cholera is one of the most widespread and damaging of such infectious microorganisms, with the World Health Organization reporting 132,000 cases in 2006 [2]. But cholera is just one of many examples. In the United States alone, there were more than 325,000 hospitalizations from food-borne illnesses in the year of 2006 [3] There are many medical treatments for food-borne illnesses, the most commonly prescribed being antibiotics. Unfortunately, antibiotics are indiscriminant and lead to rapid depletion of benign bacterial populations within the intestine. Due to this fact, dietary supplements have been popularized which aim to introduce beneficial bacteria back into the gut after antibiotic treatment; these substances have been termed ‘probiotics’. Natural probiotics have two main advantages: they provide useful functions for the host and they competitively inhibit growth of pathogenic bacteria. Natural probiotics do not convey any more advantages than bacteria in a healthy human intestine. Modern synthetic biology techniques should allow us to create an engineered probiotic that goes beyond the limitations of natural probiotics. The viability of such engineered probiotics within humans has already been established [4]. As part of a collaborative effort by the Caltech iGEM team to create a novel probiotic with improved medical applications, this project focuses on engineering a pathogen defense system within E. coli.. System DesignThe engineered probiotic acts as a delivery vehicle for phage into the large intestine. There are four important design goals for such as system: (1) the system releases phage into the large intestine, (2) the production of phage is regulated with a high dynamic range between a distinct on and off state, (3) the system can target a wide range of pathogenic bacteria, and (4) the system integrates well within the existing iGEM probiotic. An obvious delivery mechanism would be the adaptation of natural lysogens, a phase of the phage life cycle in which the phage integrates its DNA with the host organism. Lysogens lie dormant until disturbed by some external stimuli, but can be stimulated to enter the lytic cycle and release phage. There is one major limitation to this method: phage released will be specific to the lysogenic hosts, E. coli. We can avoid this limitation by incorporating as a lysogen a phage which is not normally infectious to E. coli. This phage could potentially target pathogenic bacteria besides E. coli. To demonstrate the feasibility of this approach, a simple model system was developed using λ phage and JW3996-1, a strain of E. coli immune to λ infection due to lamB deletion[5]. In this model, the host is immune to infection unless lamB is expressed on a plasmid. After infection and selection for lysogens, the lamB plasmid is counterselected against to regain immunity. Extending this model to a more realistic situation, we envision using a phage which targets a pathogenic bacterial species, constitutively expressing the phage receptor within E. coli to create lysogens, and inducing the lysogens to infect non-E. coli pathogenic bacteria.
The second plasmid which must be designed controls the induction of the lysogens to release phage into the environment. Control of this aspect of the system is vital for integration with the overall iGEM project. The induction of λ phage lysogens has been explored since 1950 [6], and serves as a model system for the study of other temperate phages. The most famous induction mechanism is ultraviolet radiation (UV). In UV induction of λ lysogens, UV induced DNA damage activates the cellular SOS mechanism, which activates the expression of RecA, a protein involved in SOS response. RecA, in turn, cleaves the λ repressor and leads to phage activation. In this study, activation of the recA pathway was not readily feasible, so an alternative effector was used to elicit induction. A secondary plasmid expressing the E. coli regulatory gene, rcsA, is used as the trigger for λ phage induction. rcsA is one of several regulatory genes which provide a recA independent pathway for λ induction[7]. Overexpression of rcsA leads to λ induction, and, furthermore, rcsA appears to directly compete with the λ suppression gene cI. [6] In this project, rcsA is used to trigger phage production. The effectiveness and dynamic range of rcsA induction is explored.
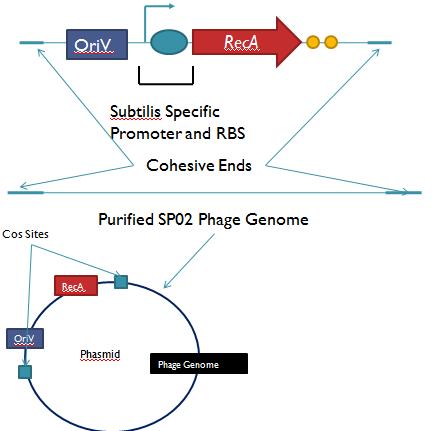
Part II: B. Subtilis LysogensBasic IdeaWe wish to create a phasmid, basically the lysogen genome with an E. Coli plasmid origin of replication, using B. Subtilis lysogens. This allows the phage genome to pass on as a plasmid within our engineered E. Coli, but when the plasmid is conjugated to B. Subtilis, the virus is induced and destroys the pathogens. Phasmid construction will an E Coli origin of replication with a Subtilis specific promoter in front of RecA or another inducer. Current ProgressThree different bacteriophage lysogens and three wildtype strains of B. Subtilis has been ordered from the Bacillus Stock Center. However, efforts to induce the bacteriophage from the lysogens with UV exposure has proved to be futile thus far, however, this was possibly due to a contamination of the lysogen stocks. The strains have been reordered and B. Subtilis lysogen induction with UV occurring soon, most likely the week of 8/4. |
 "
"