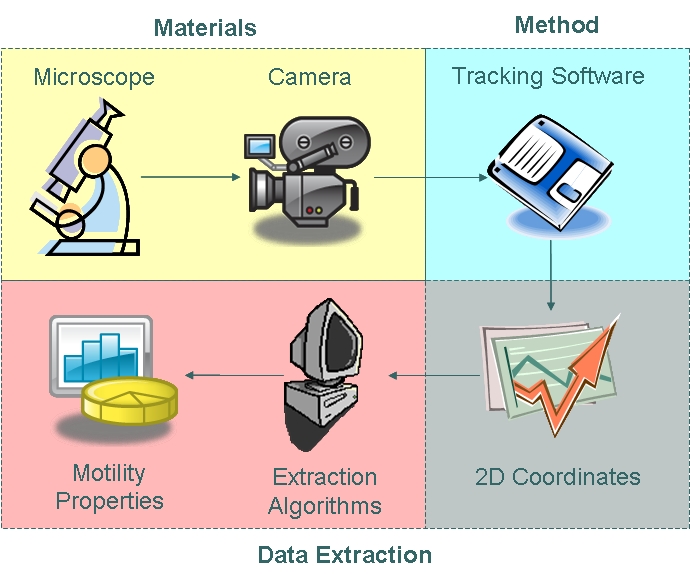
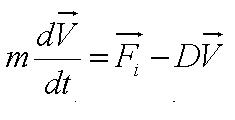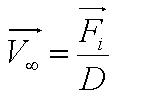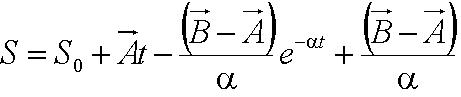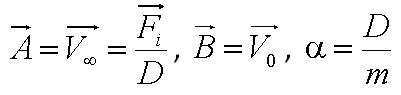Team:Imperial College/Motility
From 2008.igem.org
(Difference between revisions)
m |
|||
| Line 38: | Line 38: | ||
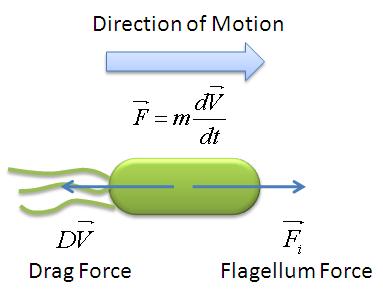
In this model, we attempt to obtain a distribution for the flagellar force, which is represented by parameter ''A''. We assume that the medium is homogenous and the drag coefficient is constant throughout the medium, hence the distribution of flagellar force will be sufficiently be described by parameter A.|}} | In this model, we attempt to obtain a distribution for the flagellar force, which is represented by parameter ''A''. We assume that the medium is homogenous and the drag coefficient is constant throughout the medium, hence the distribution of flagellar force will be sufficiently be described by parameter A.|}} | ||
| - | {{Imperial/ | + | {{Imperial/Box1|Results| |
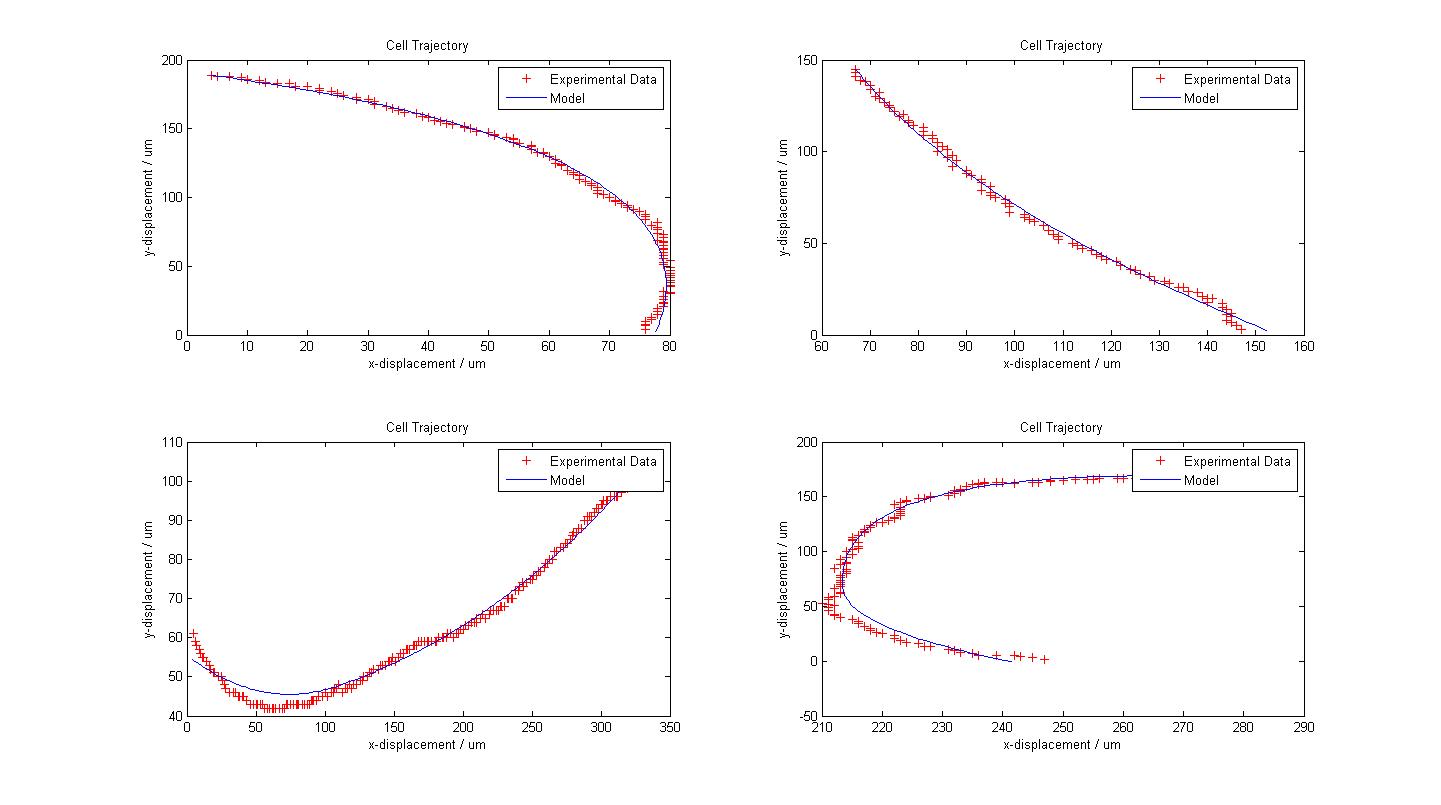
The following figure shows the results of our model fitting. We have introduced a change in flagellar force at certain points of the cell trajectory so as to achieve a better fit. A maximum of two runs were allowed for each cell trajectory. | The following figure shows the results of our model fitting. We have introduced a change in flagellar force at certain points of the cell trajectory so as to achieve a better fit. A maximum of two runs were allowed for each cell trajectory. | ||
| Line 49: | Line 49: | ||
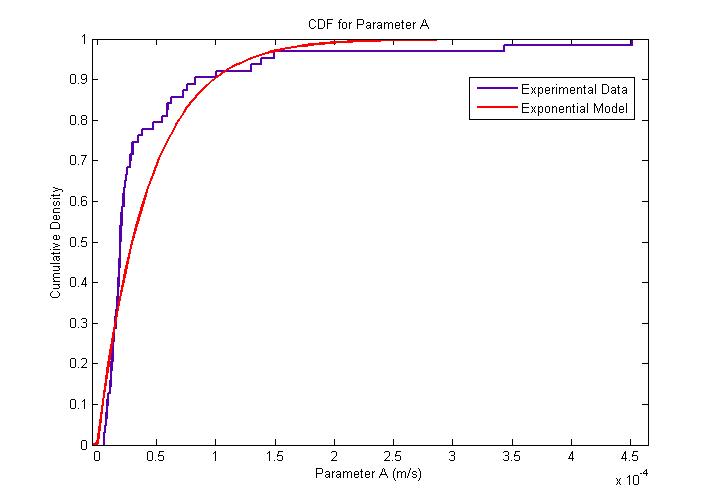
[[Image:Exponential_Distribution_for_Parameter_A_CDF.jpg|450px|center]]<br> |}} | [[Image:Exponential_Distribution_for_Parameter_A_CDF.jpg|450px|center]]<br> |}} | ||
| - | {{Imperial/ | + | {{Imperial/Box2|Conclusion| |
From our model fitting process, we can see that flagellum force is exponentially distributed. Our mechanical model though simple, fits the cell trajectory data extremely well as shown in the figure above. Further work which can be done would be to utilise a movable stage to track the movement of ''B. subtilis'' over its entire run so as to obtain a distribution of other motility parameters associated running and tumbling events. |}} | From our model fitting process, we can see that flagellum force is exponentially distributed. Our mechanical model though simple, fits the cell trajectory data extremely well as shown in the figure above. Further work which can be done would be to utilise a movable stage to track the movement of ''B. subtilis'' over its entire run so as to obtain a distribution of other motility parameters associated running and tumbling events. |}} | ||
{{Imperial/EndPage|Genetic Circuit|Appendices}} | {{Imperial/EndPage|Genetic Circuit|Appendices}} | ||
 "
"