Team:ETH Zurich/Project/Background
From 2008.igem.org
(→Top-down) |
|||
| Line 39: | Line 39: | ||
| - | [[Image:topdown.png]] | + | [[Image:topdown.png|center]] |
<!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | <!-- PUT THE PAGE CONTENT BEFORE THIS LINE. THANKS :) --> | ||
|} | |} | ||
Revision as of 20:44, 25 October 2008
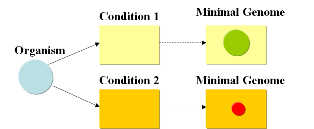
Current approachesAs usual when takling engineering projects, two main approaches can be followed: bottom-up and top-down. We start with a given set of genes and the goal is to find the subset that contains the minimal amount of genes necessary to support life under given conditions. A specific medium containing metabolites needed for growth (such as LB) will require a different minimal set of genes than a minimal medium such as M9.
A minimal genome is the minimal set of genes able to sustain life in a particular condition.
Bottom-upIn the first case we try to identify all the necessary functions for our system to work (in this case: to live). In this case we identify pathways to produce all necesary metabolites the cell needs, such as lipids, aminoacids, etc. A good example of this approach can be found in []. The following step is to sinthesize the complete chromosome with the identified genes into an "empty" cell. This approach is beeing followed e.g. by the Craig Venter Institute [].
Top-downThe second approach starts from a working syspem (such as a well characterized strain like K12). By identifying non-essential parts of the metabolism and deleting them, we reduce the complexity of the cell. Many groups are working on this method, such as the Biofrontier Laboratories [] or Scarab Genomics [].
|
 "
"