Team:Tokyo Tech/Development of low pressure inducible promoter
From 2008.igem.org
(→Strategy) |
|||
| (19 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
!align="center"|[[Team:Tokyo_Tech/Result|Result]] | !align="center"|[[Team:Tokyo_Tech/Result|Result]] | ||
!align="center"|[[Team:Tokyo_Tech/Team|The Team]] | !align="center"|[[Team:Tokyo_Tech/Team|The Team]] | ||
| - | !align="center"|[[Team:Tokyo_Tech/ | + | !align="center"|[[Team:Tokyo_Tech/Acknowledgements|Acknowledgements]] |
|} | |} | ||
| Line 14: | Line 14: | ||
--> | --> | ||
| - | + | ||
| - | + | ||
| - | + | == <font size="5">Development of low pressure inducible promoter </font>== | |
| - | + | <html> | |
| + | <body> | ||
| + | <div> | ||
| + | It is known that lac promoter is induced under 30 MPa (Kato et al. 1994).However, 30 MPa is too high to use as input. | ||
| + | </div> | ||
| + | <div> | ||
| + | Therefore, we are trying to develop low pressure inducible promoter. | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | == Method == | ||
| + | === Principle === | ||
| + | LacI binds to lacI binding site and repress lac promoter. | ||
| + | In addition, pressure of 30 MPa activated the lac promoter. | ||
| + | We propose a hypothesis that 30 MPa induce a conformational change in lacI reprssure and change the affinity for lacI binding site weaker. Therefore, lacI can't repress lac promoter under 30 MPa. | ||
| + | |||
| + | === Strategy === | ||
| + | <gallery widths="360px" heights="190px" > | ||
| + | Image:Tech dev 1.JPG|Figure 1 - RCP random mutagenesis | ||
| + | Image:Tech dev 2.JPG|Figure 2 - Strategy of sorting with FACS | ||
| + | </gallery> | ||
| + | |||
| + | We are trying to develop low pressure inducible promoter by PCR random mutagenesis to lac promoter in order to reduce affitity to lacI. | ||
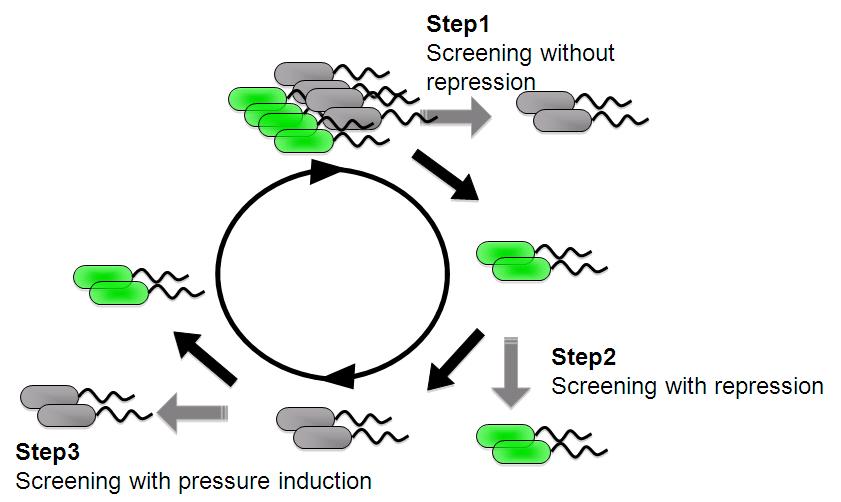
| + | And we are screening an E. coli library for promoters that are induced under low pressure using a fluorescence activated cell sorter (FACS). | ||
| + | This scheme is based on the ability to separate bacteria with a FACS in response to expression, or lack of expression, of a fluorescent marker. | ||
| + | *'''step 1''' - Fluorescent bacteria without repressor protein were collected by FACS. This sorted pool contains bacteria bearing both constitutive and low pressure-inducible promoter. | ||
| + | |||
| + | *'''step 2''' - Constitutive promoter are removed with repressor protein and sorting all non-fluorescent bacteria. | ||
| + | |||
| + | *'''step 3''' - A final passage through under pressure with repressor protain and sorting for fluorescent bacteria removes false negatives and enriches for bacteria bearing promoter that are low pressure inducible. | ||
| + | |||
| + | == Results== | ||
| + | We finished step 1. | ||
| + | Fluorescent and non-fluorescent bacteria were sorted and we characterized their promoter. | ||
| + | === Sequence and Characterization=== | ||
| + | <gallery widths="430px" heights="220px" > | ||
| + | Image:Dev 3.JPG|Figure 3 - FACS | ||
| + | Image:Tech dev 4.JPG|Figure 4 - Sequensing | ||
| + | </gallery> | ||
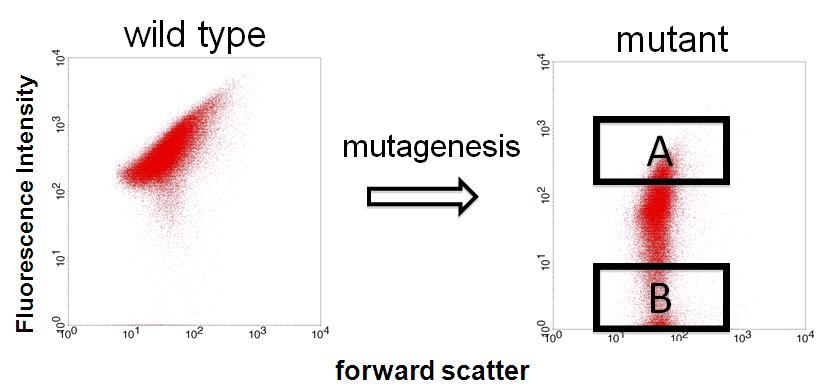
| + | We sorted fluorescent (A) and non-fluorescent bacteria (B) with a FACS. | ||
| + | Then, we analyze these base sequences. | ||
| + | *A have mutations in lacI binding site or non-functional DNA. | ||
| + | *B have mutations in CAP binding site, -35 or non-functional DNA. | ||
| + | |||
| + | == Conclusion== | ||
| + | We have successfully demonstrated that it is possible to collect objective promoter by PCR random mutagenesis and screening with a FACS. | ||
| + | So we clearly believe that we can screen low pressure inducible lac promoter mutant with this strategy. | ||
Latest revision as of 11:01, 26 October 2008
| Home | Construction | Acrylic container | Development of promoter | Genetic toggle switch | Parts Submitted to the Registry | Result | The Team | Acknowledgements |
|---|
Contents |
Development of low pressure inducible promoter
Method
Principle
LacI binds to lacI binding site and repress lac promoter. In addition, pressure of 30 MPa activated the lac promoter. We propose a hypothesis that 30 MPa induce a conformational change in lacI reprssure and change the affinity for lacI binding site weaker. Therefore, lacI can't repress lac promoter under 30 MPa.
Strategy
We are trying to develop low pressure inducible promoter by PCR random mutagenesis to lac promoter in order to reduce affitity to lacI. And we are screening an E. coli library for promoters that are induced under low pressure using a fluorescence activated cell sorter (FACS). This scheme is based on the ability to separate bacteria with a FACS in response to expression, or lack of expression, of a fluorescent marker.
- step 1 - Fluorescent bacteria without repressor protein were collected by FACS. This sorted pool contains bacteria bearing both constitutive and low pressure-inducible promoter.
- step 2 - Constitutive promoter are removed with repressor protein and sorting all non-fluorescent bacteria.
- step 3 - A final passage through under pressure with repressor protain and sorting for fluorescent bacteria removes false negatives and enriches for bacteria bearing promoter that are low pressure inducible.
Results
We finished step 1. Fluorescent and non-fluorescent bacteria were sorted and we characterized their promoter.
Sequence and Characterization
We sorted fluorescent (A) and non-fluorescent bacteria (B) with a FACS. Then, we analyze these base sequences.
- A have mutations in lacI binding site or non-functional DNA.
- B have mutations in CAP binding site, -35 or non-functional DNA.
Conclusion
We have successfully demonstrated that it is possible to collect objective promoter by PCR random mutagenesis and screening with a FACS. So we clearly believe that we can screen low pressure inducible lac promoter mutant with this strategy.
 "
"