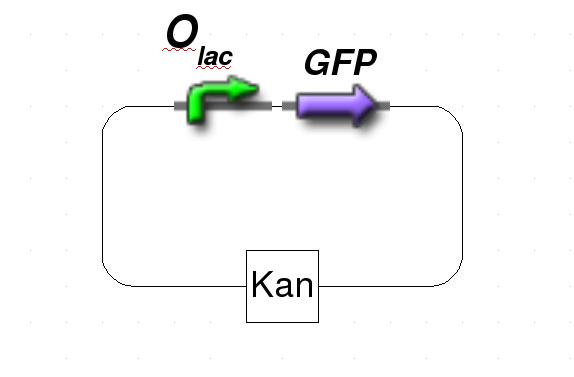
Switch Curcuit
Introduction
Many biological applications require a pulse mechanism, that enables on the one side a fast production and high accumulation of specific protein at one time and on the other side a fast elimination of this protein after a period of time has passed. Pulse generators that turn on and off the protein production are essential parts of engineered genetic circuits, which allow to control system of interest. With regard to our project, especially to the in vivo chromosomal genome-deletion, it is crucial to express restriction enzymes only for a very short amount of time in order to increase cell survival.
Recent studies describe several engineered pulse-generating circuits (1), which are based on feed-forward motif. These are parameter-sensitive and are difficult to construct biologically. We propose a novel switching mechanism which requires a start signal, realized by induction with IPTG, and a stop signal, realized by induction with tet. This setup allow us to test genetic circuit with protein pulses of different duration. Moreover, because our setup relies on standard promoters and repressor binding sites usual expression vectors such as the pET system can be used without modification.
System Description
The system is based on two plasmids. The first plasmid carry protein of interest that is under control of standard lacI repressor. The protein of interest is either fluorescent protein GFP for testing or specific restriction enzyme to support gene-deletion mechanism. In further description we will refer to GFP as a protein of interest . The second plasmid contains lacI IS, IPTG-insensitive mutant, that is under control of tet repressor and a source of consitutively expressed tetR protein. As the system will only work with both plasmids integrated, the first plasmid has Ampicillin resistance while plasmid two has Kanamycin resistance.
Mechanism
Expression of the lac operon in E. coli is tightly controlled by lacI, a protein, which binds to a repressor
binding site within the promotor and disables transcription by obscuring the promotor region. When bound to DNA, lacI is in the tetrameric form, which consists of two dimers interacting at the end
distal from the DNA binding site. Since our protein of interest, GFP is under control of lacI, there is no expression of GFP in the beginning. In order to turn of the expression of GFP the start signal, induction by IPTG, is needed. Upon binding of IPTG, the lacI tetramer breaks down into two dimers and the affinity for the repressor binding site is greatly reduced: the lacI IPTG complex will
diffuse away from the repressor binding site, leaving the promotor accessible. Now the transcription and translation of GFP protein can start.
The mechanism for termination of GFP- expression is very similar. Since lacI IS, is under tetR-repessible promoter, no lacI IS, is present. By inducing with tet, the TetR affinity for the repressor binding side is reduced and the transcription of LacI IS,can start. LacI IS on its part binds to the repressor binding side of GFP promoter which causes the termination of GFP translation.
Components Description
Here the non-standard components are described.
LacIS-mutant
LacI IS- is a protein which abolish IPTG response upon mutation.
The x-ray crystal structure of lacI with bound IPTG has allowed the identification of residues
that interact with IPTG and which are promising targets for mutagenesis (2). We decided to mutate residues R197 and T276, which are located in the IPTG binding groove, contact IPTG and have been
shown to produce the lacI IS mutation in previous genetic experiments. We decided to generate a set of eight mutated lacIs, in which we replaced either R197 with alanine or phenylalanine or T276 with alanine or phenylalanine or both in all possible combinations.
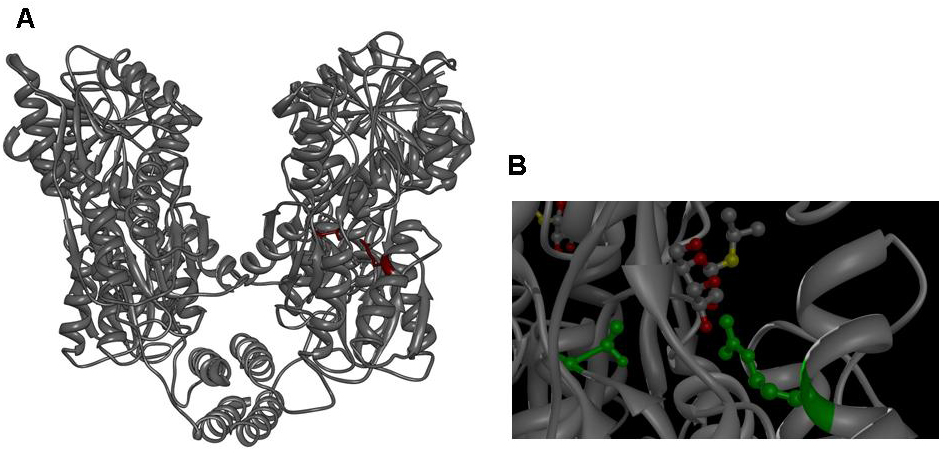 Figure 1: A Lac repressor tetramer, residues R197 and T276 are shown in red. B IPTG bound to the inducer binding site of the lac repressor, residues R197 and T276 are shown in green. Molecular graphics was generated from coordinate set [http://www.rcsb.org/pdb/explore.do?structureId=1LBH 1lbh] (27) with [http://www.cgl.ucsf.edu/chimera/ UCSF Chimera].
|
 "
"