Team:NTU-Singapore/Parts/New Proposal
From 2008.igem.org
Lalala8585 (Talk | contribs) |
Lalala8585 (Talk | contribs) |
||
| (5 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
| - | = | + | ='''Proposed New Characterization protocol'''= |
| - | An important part of the iGEM project is to perform characterization of Biobricks. | + | An important part of the iGEM project is to perform characterization of Biobricks. For the desired promoter to be characterized a GFP gene is attached behind it and the resulting fluorescence is measured under different conditions. However, after reviewing the different websites and protocols from different teams, we realized that there are a myriad of methods out there. Furthermore, one of the problems faced was that each team may use different equipments to measure the fluorescence readings. These readings are measured against a reference that is different for different equipments used. |
| - | + | Hence we proposed a new approach, which measures the protein concentration via SDS page. Our method would allow different teams to perform their own characterization experiments and still be able to share their findings in a more methodical way. The same machines could be used to measure the fluorescence units. But they would have to be cross-referenced with a standard SDS-page run that could quantity the concentration of protein formed. | |
| - | + | ||
| - | In an attempt to quantify the concentration of GFP protein formed relative to the RFU measurements, we took cell samples of varying RFU values and store the cells in | + | Therefore the fluorescence readings would have a physical meaning of equivalent amount of protein formed. |
| + | |||
| + | =='''Initial Concept'''== | ||
| + | ===Quantifying the concentration of Protein formed=== | ||
| + | |||
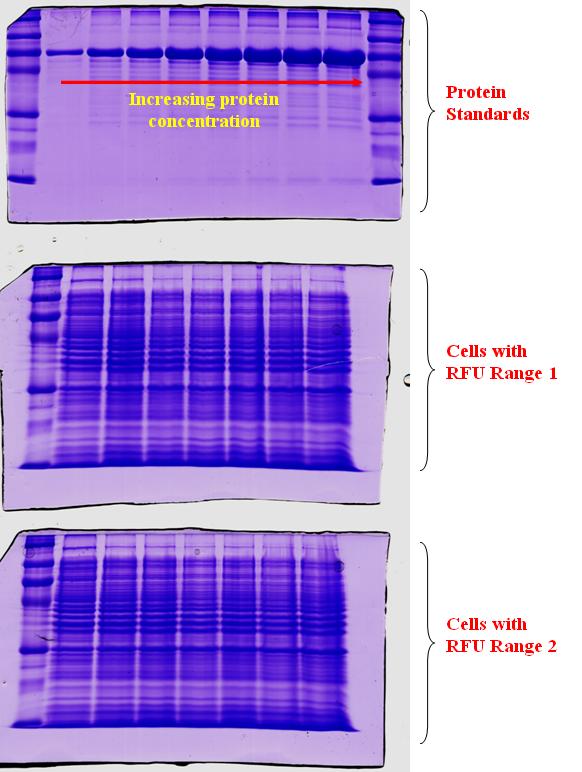
| + | In an attempt to quantify the concentration of GFP protein formed relative to the RFU measurements, we took cell samples of varying RFU values and store the cells in -20°C till a corresponding range of RFU values were collected. After which, varying protein standards of known concentration were prepared and a gel run was conducted as depicted in the top gel run of the diagram below. As shown in the gel run, with an increasing protein concentration, the thickness of the band would also increase. The next two gel runs in the diagram were for the range of RFU investigated. At higher RFU readings, we would expect a higher protein concentration present. Hence, a thicker band should also be seen. However, for the increasing range of RFU readings, we do not obtain an expected increase in thickness of the band. | ||
This could be due to the following reasons<br> | This could be due to the following reasons<br> | ||
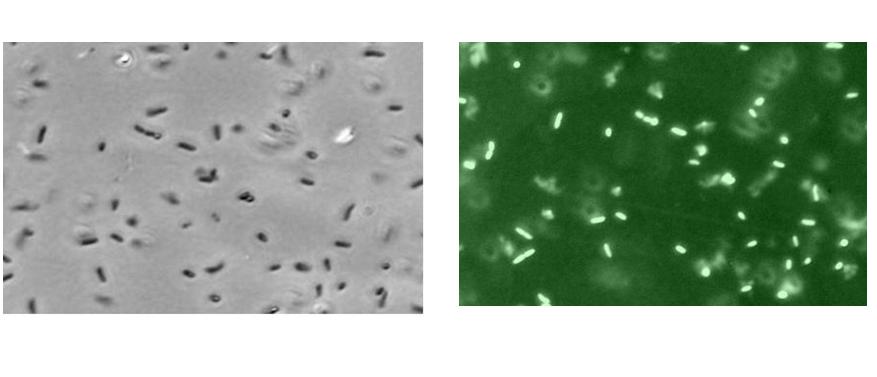
| - | 1) The increase in RFU was because of the increase in cell number and not the increase in concentration of the GFP protein.<br> | + | 1) The increase in RFU was because of the increase in cell number and not the increase in concentration of the GFP protein. But we managed to prove that this was not true, as we took electron microscope pictures and showed that cells did actually glow brighter. |
| + | |||
| + | [[Image:NTU@iGEM_Fluorscence_LacI-GFP_1mM.JPG|thumb|center|400px|Fluorescence Microscope picture of pLacI-GFP at 1.0mM Lactose concentration]] | ||
| + | |||
| + | |||
| + | [[Image:NTU@iGEM_Fluorscence_LacI-GFP_10mM.JPG|thumb|center|400px|Fluorescence Microscope picture of pLacI-GFP at 10.0mM Lactose concentration]] | ||
| + | |||
| + | <br> | ||
2) The increase in band thickness was not distinctive, as the concentration GFP protein when compared to the other proteins in the cell may be low.<br> | 2) The increase in band thickness was not distinctive, as the concentration GFP protein when compared to the other proteins in the cell may be low.<br> | ||
| Line 21: | Line 32: | ||
The experiment we carried out was to obtain cells inoculated with lactose during the characterization process and to extract the protein from them. The RFU recorded at a particular time would be correlated with the amount of protein that the cell had within during the extraction. The protein quantification would come from a gel run of the proteins against a standard ladder. | The experiment we carried out was to obtain cells inoculated with lactose during the characterization process and to extract the protein from them. The RFU recorded at a particular time would be correlated with the amount of protein that the cell had within during the extraction. The protein quantification would come from a gel run of the proteins against a standard ladder. | ||
| - | |||
| - | |||
| + | =='''Results'''== | ||
| + | The experiment showed that the amount of proteins in the cells was alike. | ||
Since the experiment did not yield the expected results, it was possible that the lactose provided the cells with the nutrients to multiply. A higher lactose concentration could mean that the cells were able to grow at a faster rate and produce a higher fluorescence reading. | Since the experiment did not yield the expected results, it was possible that the lactose provided the cells with the nutrients to multiply. A higher lactose concentration could mean that the cells were able to grow at a faster rate and produce a higher fluorescence reading. | ||
What this means for us is that the increase in RFU(flouresence)could be linked more closely to the ability for the cell to reproduce under different lactose inoculations, and the models which describe faster or better protein production itself could be flawed. | What this means for us is that the increase in RFU(flouresence)could be linked more closely to the ability for the cell to reproduce under different lactose inoculations, and the models which describe faster or better protein production itself could be flawed. | ||
| + | <br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| + | |||
| + | var tags_before_clock = "<b>It is now " | ||
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.write('<layer id="clock"></layer><br>'); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | document.write('<span id="clock"></span>'); | ||
| + | } | ||
| + | |||
| + | DaysofWeek = new Array() | ||
| + | DaysofWeek[0]="Sunday" | ||
| + | DaysofWeek[1]="Monday" | ||
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| + | |||
| + | Months = new Array() | ||
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | setInterval("upclock()",1000); | ||
| + | //--> | ||
| + | </script> | ||
| + | </html> | ||
Latest revision as of 05:51, 27 October 2008
|
Contents |
Proposed New Characterization protocol
An important part of the iGEM project is to perform characterization of Biobricks. For the desired promoter to be characterized a GFP gene is attached behind it and the resulting fluorescence is measured under different conditions. However, after reviewing the different websites and protocols from different teams, we realized that there are a myriad of methods out there. Furthermore, one of the problems faced was that each team may use different equipments to measure the fluorescence readings. These readings are measured against a reference that is different for different equipments used.
Hence we proposed a new approach, which measures the protein concentration via SDS page. Our method would allow different teams to perform their own characterization experiments and still be able to share their findings in a more methodical way. The same machines could be used to measure the fluorescence units. But they would have to be cross-referenced with a standard SDS-page run that could quantity the concentration of protein formed.
Therefore the fluorescence readings would have a physical meaning of equivalent amount of protein formed.
Initial Concept
Quantifying the concentration of Protein formed
In an attempt to quantify the concentration of GFP protein formed relative to the RFU measurements, we took cell samples of varying RFU values and store the cells in -20°C till a corresponding range of RFU values were collected. After which, varying protein standards of known concentration were prepared and a gel run was conducted as depicted in the top gel run of the diagram below. As shown in the gel run, with an increasing protein concentration, the thickness of the band would also increase. The next two gel runs in the diagram were for the range of RFU investigated. At higher RFU readings, we would expect a higher protein concentration present. Hence, a thicker band should also be seen. However, for the increasing range of RFU readings, we do not obtain an expected increase in thickness of the band.
This could be due to the following reasons
1) The increase in RFU was because of the increase in cell number and not the increase in concentration of the GFP protein. But we managed to prove that this was not true, as we took electron microscope pictures and showed that cells did actually glow brighter.
2) The increase in band thickness was not distinctive, as the concentration GFP protein when compared to the other proteins in the cell may be low.
The experiment we carried out was to obtain cells inoculated with lactose during the characterization process and to extract the protein from them. The RFU recorded at a particular time would be correlated with the amount of protein that the cell had within during the extraction. The protein quantification would come from a gel run of the proteins against a standard ladder.
Results
The experiment showed that the amount of proteins in the cells was alike. Since the experiment did not yield the expected results, it was possible that the lactose provided the cells with the nutrients to multiply. A higher lactose concentration could mean that the cells were able to grow at a faster rate and produce a higher fluorescence reading.
What this means for us is that the increase in RFU(flouresence)could be linked more closely to the ability for the cell to reproduce under different lactose inoculations, and the models which describe faster or better protein production itself could be flawed.
 "
"