Team:ETH Zurich/Project/Background
From 2008.igem.org
(→Top-down) |
m |
||
| Line 4: | Line 4: | ||
</center> | </center> | ||
</html> | </html> | ||
| - | |||
{|style="background:#C6E2FF ; border:3.5px solid #60AFFE; padding: 1em; margin: auto ; width:98.5% " | {|style="background:#C6E2FF ; border:3.5px solid #60AFFE; padding: 1em; margin: auto ; width:98.5% " | ||
|- | |- | ||
Revision as of 19:59, 27 October 2008
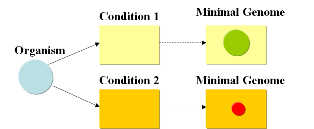
Current approachesAs usual when tackling engineering projects, two main approaches can be followed: bottom-up and top-down. We start with a given set of genes and the goal is to find the subset that contains the minimal amount of genes necessary to support life under given conditions. A specific medium containing metabolites needed for growth (such as LB) will require a different minimal set of genes than a minimal medium such as M9. This is due to the fact, that a cell living under conditions where metabolites are missing, will require additional genes that encode for the supplemental enzymes for biosynthesis and energy production in order to able to sustain life (grow and reproduce). It will also need to have the possibility to adapt, if different sources of energy are available at different times.
A minimal genome is the minimal set of genes able to sustain life in a particular condition.
Bottom-upIn the first case we try to identify all the necessary functions for our system to work (in this case: to live). In this case we identify pathways to produce all necesary metabolites the cell needs, such as lipids, aminoacids, etc. A good example of this approach can be found in []. Here the chromosome of Mycoplasma genitalium with 580 kbp and 482 protein-coding genes has been selected as a starting point. Further analysis of the required functions has led to the selection of 382 genes in the chromosome. The following step is to synthesize the complete chromosome with the identified genes and transfer it into an "empty" cell (this approach is being followed e.g. by the JCVI []). Top-downThe second approach starts from a working system (such as a well characterized strain like K12). By identifying non-essential parts of the metabolism and deleting them, we reduce the complexity of the cell. Many groups are working on this method, such as the Biofrontier Laboratories [] or Scarab Genomics [].
|
 "
"