Team:Cambridge/Bacillus/Lab Work
From 2008.igem.org
(→August 1st) |
(→August 5th) |
||
| Line 807: | Line 807: | ||
- ECE176 replated onto Amp100 + Cm5 | - ECE176 replated onto Amp100 + Cm5 | ||
| + | |||
| + | |||
| + | =August 6th= | ||
| + | |||
| + | ==PCR parts of Agr operon== | ||
| + | |||
| + | We received primers for Agr A,B,C, and D today - http://openwetware.org/wiki/IGEM:Cambridge/2008/Turing_Pattern_Formation/Primers | ||
| + | |||
| + | We must PCR each individual part out of the previous BioBricks (I746001 &I746101) in order to put our bacillus RBS on them. | ||
| + | |||
| + | I was concerned that the region of most homology would be the BioBrick prefix and suffix itself, so I first digested the DNA with Xbal and SpeI. | ||
| + | |||
| + | For some reason the digestion didn't work that well, so I've decided to go ahead and use the uncut BioBrick DNA as template for my reactions. | ||
| + | |||
| + | |||
| + | =August 9th= | ||
| + | |||
| + | ==Plans for Monday== | ||
| + | ===Lab Work=== | ||
| + | *Be sure that the Spc50 plates are working, as the control for Spc50 grew and shouldn't have. We already know that things can die at Spc100 (the double crossover test from earlier this week) so it is unlikely that the stock is bad. We should test Spc50 again with regular bacillus and with E. coli at 50-100 ug/ml and hope that they die. | ||
| + | *Finish all the agr PCRs, but especially the RBS. | ||
| + | *Confirm that amyE insertion works, using the amylase/ery tests. | ||
| + | *Confirm that with ECE153 inserted, we get detectable fluoresence in a microscope. | ||
| + | *Confirm that ECE166 transformation works, using plasmid miniprepping. | ||
| + | **Find the qiagen miniprep kit or adapt the protocol. | ||
| + | *Confirm that when ECE166 is transformed, we get detectable fluoresence in a microscope. | ||
| + | ===Book/Computer Work=== | ||
| + | *Find good promoters in vectors and order PCR primers to get them out | ||
| + | *Find out how to assay for/make AIP | ||
| + | **Find/Contact Jim Ajioka | ||
| + | *Find out how to do a c-terminal GFP fusion for membrane proteins agrB and agrC | ||
| + | **Can we fuse GFP and still expect them to localize to the membrane? | ||
| + | ***AgrB transmembrane topology (http://www.jbc.org/cgi/content/full/277/38/34736#F1). The signal peptide is not on the N terminus, and both the N and C terminus are intracellular. | ||
| + | ***AgrC transmembrane topology (http://openurl.ingenta.com/content?genre=article&issn=0950-382X&volume=28&issue=3&spage=655&epage=662 (see figure 2 B)), the N terminus is probably extracellular. The C terminus is probably intracellular. The article doesn't say where the signal peptide is. | ||
| + | **Should we do a antibody tag instead? | ||
| + | |||
| + | ==Longer Term Plans== | ||
| + | *Ligate the RBS into a BB vector so we can build a RBS/GFP construct. | ||
| + | *Put the RBS/GFP construct into ECE166 and ECE 153. | ||
| + | *Compare the 'out of the box' ECE153 with ECE153 with RBS/GFP biobrick inserts | ||
| + | *Compare the 'out of the box' ECE166 with ECE166 with RBS/GFP biobrick inserts | ||
| + | |||
| + | |||
| + | =August 11th= | ||
| + | ==PCR Bacillus RBS== | ||
| + | |||
| + | We created 2 different Bacillus RBS sites simply by primer annealing and extension. Each product includes an 8bp RBS plus the BioBrick prefix and suffix for a total of 54bp. | ||
| + | The products were visualized on a 3.5% Agarose gel with bioline's hyperladder V. | ||
| + | |||
| + | RBSs is the consensus RBS sequence for Bacillus subtilis, and thus we believe it to be a very strong binding site - AAAGGAGG | ||
| + | |||
| + | RBSw has a 2bp modification from RBSs, which we believe will weaken it - AGAGGTGG | ||
| + | |||
| + | |||
| + | The amplification of RBSs yielded only one band on the gel. The product was purified using microclean and contained 14.8ng/uL after clean-up. | ||
| + | |||
| + | RBSw, however produced 2 bands on the gel. The correct size was gel-extracted and purified. After purification the yield was 12.3 ng/uL. | ||
| + | |||
| + | |||
| + | =August 13th= | ||
| + | |||
| + | ==PCR Agr A, B, C, D== | ||
| + | |||
| + | |||
| + | Doing a PCR directly from the old Biobricks has worked for all parts of the operon. No prior digestion seems to be needed. | ||
| + | |||
| + | {|class="wikitable" style="text-align:center" border="1" | ||
| + | |- | ||
| + | ! Part !! 260/280 !! μg/mL | ||
| + | |- | ||
| + | | A || 1.78 || 92.5 | ||
| + | |- | ||
| + | | B || 1.87|| 96.5 | ||
| + | |- | ||
| + | | C || 1.9 || 127.7 | ||
| + | |- | ||
| + | | D || 1.82 || 26.3 | ||
| + | |} | ||
| + | |||
| + | |||
| + | =August 15th= | ||
| + | |||
| + | ==PCRing out promoters== | ||
| + | |||
| + | - 4 different promoters : Pxyl, Pspc, Ppac, Pupp | ||
| + | |||
| + | - Make master mix enough for 4RXNs | ||
| + | |||
| + | |||
| + | |||
| + | ==Digest and Ligate Agr A and C to pSB4C5== | ||
| + | |||
| + | Previous PCR products of Agr A and C were digested using Fermentas Fast Digest - EcoRI and PstI according to their protocol except a longer time was given to digest. | ||
| + | |||
| + | 3uL of AgrA was used and 4.4 uL of AgrC was used and incubated for 40minutes. | ||
| + | |||
| + | 10uL totaling to 1ug of pSB4C5 plasmid was also digested and incubated for 10 minutes. | ||
| + | |||
| + | The digestion was visualized on a 0.8% Agarose gel and produced expected sizes: | ||
| + | |||
| + | Plasmid: approx 3bk | ||
| + | |||
| + | Agr A: approx 700bp | ||
| + | |||
| + | Agr C: approx 1300bp | ||
| + | |||
| + | |||
| + | =August 16th= | ||
| + | |||
| + | ==Agr A and C ligation to pSB4C5== | ||
| + | |||
| + | Ligation was performed using Fermentas Rapid Ligation kit according to their protocol. | ||
| + | |||
| + | After ligation, samples were visualized on a gel, but little product of the correct size was seen. | ||
| + | |||
| + | For agrA we should have gotten a band of size 3700 (3000plasmid + 700 insert) | ||
| + | |||
| + | For agrC we should have gotten a band of size 4300 (3000plasmid + 1300insert) | ||
| + | |||
| + | |||
| + | The bands of appropriate sizes were extracted for transformation. | ||
| + | |||
| + | |||
| + | =August 18th= | ||
| + | |||
| + | ==Check promoters== | ||
| + | |||
| + | On Friday, PCR out promoters, we want to check them. | ||
| + | |||
| + | - Run a gel | ||
| + | |||
| + | - Results : good size of bands!!! | ||
| + | |||
| + | ==Transformation of AgrA and AgrC== | ||
| + | |||
| + | AgrA and C gell-extracted ligation was transformed into Top 10 cells using standard protocol. Puc9 was used as a positive control. | ||
| + | |||
Revision as of 18:13, 28 October 2008
July 22nd
We have 4 tubes from last year. These strains are frozen in glycerol.
- PNZ8901 plasmid in E.coli MC1061 Strain
- Chloroamphenicol resistant
- "sure" vector
- does not integrate
- B. subtilis strain 1A1
- No resistance
- Same as strain 168 but tryptophan deficient
- E.coli strain MC1061, no plasmid
- PP182 plasmid in E.coli strain DH5alpha
- Ampicillin resistance
- integrates
Checking antibiotic resistance
Purpose : Grow each of them on a plate to test their antibiotic resistance
Protocol
- Warm up frozen tubes
- Take 15uL of each and put it on plates without antibiotic
- Incubate at 37°C
July 23rd
- Take one colony from each of the plates grown from the tubes yesterday and plated them again on LA without antibiotics
1/ Purpose Check antibiotic resistance of our different strains
| Strain | Plasmid | Extracted from | Antibiotic use | Concentration of antibiotic |
|---|---|---|---|---|
| DH5alpha | PP182 | tube | Amp | 100 |
| MC1061 | NONE | tube | Cm | 35 |
| MC1061 | PNZ8901 | tube | Cm | 35 |
| DH5alpha | PP182 | plate | Amp | 100 |
| MC1061 | NONE | plate | Cm | 35 |
| MC1061 | PNZ8901 | plate | Cm | 35 |
2/ Purpose : Find the right concentration of antibiotic so that B. subtilis survive
- Grow 15uL of B. 1A1 (frozen tube) in 5mL of LB
- Incubate at 37°C
3/ Purpose : Grow plasmids in TOP10, transformation
4 plasmids :
- I746000
- I746100
- I746101
- I746001
1 control : PUC19
- Add 20uL of TOP10 competent cells and 0.5uL of Plasmid in an eppendorf
- 30min on ice
- Heat shock : 60s at 42°C
- 2min on ice
- Add 89.5uL of SOC
- 60min at 37°C
- Put the mix on plate with ampicillin resistance
- Incubate at 37°C
July 24th
- Test of antibiotic resistances of strains of last year
| Strain | Plasmid | Extracted from | Antibiotic use | Observations | Conclusion |
|---|---|---|---|---|---|
| DH5alpha | PP182 | tube | Amp | Many colonies | Amp Resistance OK |
| DH5alpha | PP182 | plate | Amp | Many colonies | Idem |
| MC1061 | NONE | tube | Cm | Nothing | As expecting, no Cm Resist. |
| MC1061 | NONE | plate | Cm | Nothing | Idem |
| MC1061 | PNZ8901 | plate | Cm | Maybe a few colonies | Contamination?? |
| MC1061 | PNZ8901 | tube | Cm | No colonies | no Cm Resist. |
- Transformation of E.coli with different plasmids from last year
| Strain | Inserted plasmid | Antibiotic | Observation | Conclusion |
|---|---|---|---|---|
| E.coli | I746000 | Amp | No colonies | Antibiotic resistance unknown, no Amp Resist., Pb : no terminator |
| E.coli | I746100 | Amp | No colonies | Antibiotic resistance unknown, no Amp Resist., Pb : no terminator |
| E.coli | I746001 | Amp | Many colonies | Transformation OK |
| E.coli | I746101 | Amp | Many colonies | Transformation OK |
Antibiotic test for Bacillus resistance
We want to find the lowest concentration of antibiotic which kills Bacillus.
Dilution of Amp. [ 1:1000 means 1 part stock to 1000 part Sterile Distilled Water ]
| Concentration (μg/mL) | 100 | 75 | 50 | 25 | 10 |
|---|---|---|---|---|---|
| 100 mg/mL Stock | 1:1000 | 3:4000 | 1:2000 | 1:4000 | 1:10000 |
Dilution of Cm. [ 1:1000 means 1 part stock to 1000 part Sterile Distilled Water ]
| Concentration (μg/mL) | 35 | 25 | 15 | 10 | 5 |
|---|---|---|---|---|---|
| 100 mg/mL Stock | 1:1000 | 1:1400 | 3:7000 | 1:3500 | 1:7000 |
- Add Disks in antibiotic solution for 5 mins [ Protocol to sterilize tweezers : Wipe with Kimwipes, Ethanol, Flame ]
- Melt Soft Agar
- 3mL SA + 10μL Cells 1A1 from LB prepared on 23/7/08
- Pour SA+Cells over blank hard agar plates
- put disks with different concentration of Amp/Cm above SA
- Incubate at 37°C
Results [Plates were prepared before lunch, at 5pm there were visible growth]
-Amp Plates: Huge rings of no growth around 100, 75, 50, 25, and 10.
-Cm PLates: Tiny rings of no growth around 5, 10, 15 and 25. Small ring of no growth around 35 but not well defined. (No clear zones)
Salts for making Bacillus Competent
10x Medium A Base
- Yeast Extract 10g
- Casamino Acid 2g
- add Distilled water to 900mL
-Aliquot into 5 different bottles. [180mL each]
10x Bacillus Salts
- NH4)2SO4 20g
- K2PO4 anhydrous 139.66g
- KH2PO4 60g
- Tri-Sodium Citrate 10g
- MgSO4.7H2O 2g
- Add Distilled water to 1000mL
-Aliguot into 5 different bottles. [200mL each]
Medium B
- Preparing 50mM CaCl2.2H2O
- CaCl2.2H2O 1.470g
- Add Distilled water to 20mL [Final conc. 500mM]
- Take 10mL of 500mM
- Add 90mL of distilled water [Final conc. 50mM]
- Preparing 50mM CaCl2.2H2O
- Preparing 250nM MgCl2.6H2O
- MgCl2.6H2O 0.508g
- Add Distilled water to 10mL [Final conc. 250mM]
- Take 1mL of 250mM
- Add 99mL of Distilled water [Final conc. 250μM]
- Take 1mL of 250μM
- Add 99mL of Distilled water [Final conc. 250nM]
- Preparing 250nM MgCl2.6H2O
- Add 100mL of 50mM CaCl2.2H2O
- Add 100mL of 250nM MgCl2.6H2O
-Total volume 200mL
NOTE: To complete Medium B, Take 0.2mL of this solution
July 25th
Results from Yesterday
- Antibiotic test
Results for Amp
Even with a concentration of 10μg/mL, Amp kills Bacillus. Since we want to know which is the lowest concentration of Amp which kills B.S, we are going to test with some lower concentrations.
Results for Cm
Even with a concentration of 35μg/mL, ther is not a clear area around the antibiotic disk. So we have to test some higher concentration of Cm.
Wet Work
- New antibiotic tests
Dilution of Amp
| Concentration (μg/mL) | 10 | 7.5 | 5 | 2.5 | 1 |
|---|---|---|---|---|---|
| 10 μg/mL Stock (from yesterday) | 1:1 | 3:4 | 1:2 | 1:4 | 1:10 |
Dilution of Cm
| Concentration (μg/mL) | 35 | 37.5 | 40 | 42.5 | 45 |
|---|---|---|---|---|---|
| 35 μg/mL Stock (from yesterday) | 1:1 | 5:6 (from 45μg/mL) | 1:875 | 17:18 (from 45μg/mL) | 9:7000 |
- Melt Soft Agar - 3mL SA + 10μL Cells 1A1 from LB prepared on 23/7/08 - Pour SA+Cells over blank hard agar plates - Put 5 blank disks on each agar plate - Add 40μL of antibiotic on each disk - Incubate at 37°C
Results
Nothing! 1A1 cells were kept in the freedge! B.S. can not be kept in the fridge, low temperatures kill them!
July 28th
- Results of antibiotic plates from yesterday
For Cm, 35μg/mL should be enough to kill B.S.
For Amp, nothing can be concluded!
Wet Work
- Check plasmid ppL82
We had 2 samples of ppL82 in DH5α in LB solution (~4mL), one from the frozen glycerol tube and one from a colony picked on a plate. Normally plasmid and cells should not be kept in freezer. So, we want to extract this plasmid, check its size and then keep it in the fridge. We do that for the 2 different samples.
- Plasmid Miniprep (standard protocol)
- Test the concentration of DNA in each tube
- ppL82 plate : 89.8ng/mL
- ppL82 tube : 71ng/mL
- Preparation of different stocks of strains and plasmids
PNZ8901
- Plate a single colony (from 23/07/08) on a Cm plate
- Incubate at 37°C
- Grow the entire frozen glycerol tube in 20mL of LB without antibiotic to check if this stock is still good (we will check the plasmid on a gel tomorrow)
- Incubate at 37°C
MC1061
-Pick e single colony of MC1061 (from 22/07/08) and put it in 10mL of LB (to make glycerol stocks)
-Incubate at 37°C
1A1
- Add 100μL of LB+1A (mix of friday) and 10mL of LB
- Incubate at 37°C
- Make B.S. 1A1 competent and transform them
- Prepare Medium A
Add 81mL of SDW, 10mL of 10X Medium A base and 9mL of 10X Bacillus salts
- Make B.S. 1A1 competent
- In 10mL of medium A and add about 10 colonies of B.S.
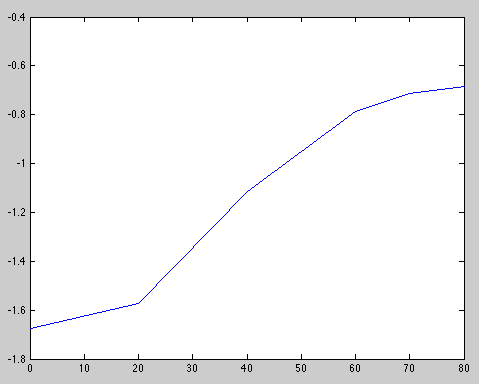
- Check the OD650, you should have an OD between 0.1 and 0.2. t0 for OD = 0.1876
- Check the OD650 every 20min and plot OD against time on semi-log paper. When the point at the culture leaves log growth, it is ok
| Time (min) | 0 | 20 | 40 | 60 | 70 | 80 |
|---|---|---|---|---|---|---|
| OD650 | 0.1876 | 0.2074 | 0.3282 | 0.4545 | 0.4895 | 0.5040 |
At, t0 = 70min, log growth seems to stop.
- Incubate at 37°C 90min after t0
- Warn 10 Ependorf tubes with 0.45mL of Medium B
- Add 50μL of culture in each tube of Medium B at t90
- Incubate the diluted culture at 37°C with vigorous aeration for 90min
- Storage : we want to store 7 tubes
6 tubes in the freezer (with 60μL of glycerol)
1 tube on the bench
- Transformation
- We make ppL82 from plate, ppL82 from tube, and a control
- Add 0.5μg of DNA (15μL)
- Incubate at 37°C for 30min
- Plate on Cm plates
July 29th
- Result from the gel (29/07/2008)
- Lane8 : ppL82 (plate)
- Lane9 : ppL82 (tube)
- Lane 10 : I746001
- Lane 11 : I746101
- Lane 12 : hyperladderI
The ladder seems to be wrong. So it was really difficult to ckeck the size of our plasmid ppL82. To make, that, we assume that the size of our biobricks was ok, and we estimate the size of our plasmid. The plasmid ppL82 seems to have the right size, but we will have to check again to be able to use a ladder.
- Competence of B.S. kept on the bench
We observed B.S. with a microscope. Problem, all B.S. seems to be dead!
Possible reasons of this problem :
- Problem with the vector (it could be a wrong vector, we are not really sure)
- Quantity of DNA, we added 0.5μg. The protocol was 1μg, but normally it should be ok.
- B.S. may not be competent (we have to test motility with microscope)
- Cells could be dead : replate them
- Try to add tryptophan in the medium
Probable reason : we forgot to dilute the medium B, that's why it did not work
- Results from transformation plates (B.S.)
There are 2 colonies on a plate, but it does not look like Bacillus. Moreover, there is also a colony on our negative control plate. SO it must be contamination (resistant to CM!). We will do the transformation again.
Wet Work
- Prepare medium for transformation of B.S.
- Preparation of medium B: 10mL of medium A and 0.2mL of 50mMCaCl22(H2O) + 250mMgCl26(H2O)
- Preparation of medium A with tryptophan : 81mL of SDW, 9mL of 10X Bacillus salts, 10mL of 10X Medium A base and 0.1mL of Tryptophan (11mg/mL)
- Check plasmid PNZ8901
- Plasmid miniprep (same protocol with 60μL of elution buffer)
- Digest : with PstI and SalI from Biolabs, and Buffer 3 (add 15μL of DNA)
- Gel (17μL of sample and 3μL of dye)
Results from the gel
- Lane9 : PNZ8901
- Lane10 : HyperladderI
The sizes we expected were about 1100kb and 2100kb. The sizes should be ok!
July 30th
- Transforming Bacillus Subtilis with medium A and medium A + tryptophan
Medium A
| Time (min) | 0 | 20 | 40 | 60 | 73 | 85 | 95 | 105 |
|---|---|---|---|---|---|---|---|---|
| OD650 | 0.1394 | 0.1370 | 0.1660 | 0.2304 | 0.2778 | 0.3138 | 0.3373 | 0.3583 |
For this medium, the curb stop to increase logarithmically at 100min.
Medium A + tryptophan
| Time (min) | 0 | 20 | 40 | 60 | 73 | 85 | 95 |
|---|---|---|---|---|---|---|---|
| OD650 | 0.1493 | 0.1636 | 0.2080 | 0.2909 | 0.3512 | 0.4002 | 0.4165 |
For this medium, the curb stop to increase logarithmically at 90min.
- Incubate 90min at 37°C by shaking
- Prepare 7 tubes for medium A and 7 tubes for medium A + tryptophan : add 0.45mL of prewarmed medium B and 0.05mL of culture and incubate 90min at 37°C by shaking.
- Transform B.S. by adding DNA and incubate 30min at 37°C
We have one transformation with ppL82 (add 7.5μL of DNA), one with PNZ8901 (add 10μL of DNA) and one negative control without DNA.
- Plate 500μL on Cm plate (35μg/mL)
- For the remaining tubes, in one, add 60μL of glycerol and keep it in the freezer and keep the other one on the bench (for each medium)
- Checking our big stock of biobricks and PNZ plasmid
We did big culture of biobricks and PNZ8901 plasmid to be able to make stocks. We want to check them before making stocks.
- Plamsid miniprep (for I746001, I746101 and PNZ8901 from big flasks
- Measure DNA concentration in our samples to decide the volume of DNA we have to add in our preparation
| 260/280 | DNA concentration (ng/nl) | |
|---|---|---|
| I746001 | 1.79 | 50.5 |
| I746101 | 1.87 | 54.3 |
| PNZ8901 | 1.84 | 87 |
- Digest
| For I746001 and I746101 | For PNZ8901 plasmid |
|---|---|
| 14μL of SDW | 14μL of SDW |
| 2μL of 10X Fast Digest Buffer | 2μL of Buffer 3 (Biolabs) |
| 2μL of DNA of biobricks | 2μL of DNA of PNZ8901 |
| 1μL of EcoRI | 1μL of PstI |
| 1μL of SpeI | 1μL of SalI |
- Gel
- Results from this gel
- Lane 9 : I746001
- Lane 10 : I746101
- Lane 11 : PNZ8901
- Lane 12 : HyperladderI
Everthing is really too big! There is a problem, either dimerization, either contamination, either a problem in our work. So we are going to run a new gel.
- New gel to check
- Miniprep plasmid from growth bottles (I746001, I746101 and PNZ8901); from plates (I746001, I746101 and PNZ8901).
- Do single digest for PNZ8901 (one with PstI, one with SalI)
- Run a gel with : PNZ8901 from friday, PNZ8901 digest with PstI, PNZ8901 digest with SalI, 3 samples from growth bottles, 3 samples from plates (to check the size of the uncut vectors)
- Result from this second gel
The ladders are really bad! But the size of our biobricks and plasmid are too big! So we can not trust these big cultures. They is a problem. With tests from the first week, we know that biobricks are right, so we are going to grow new cultures from these first cultures, to make stocks.
Concerning the vectors, they are from last year, so we are not sure of what they are. Since we ordered new well defined vectors (we should receive them on friday), we will use them in the next steps to be sure of our work.
August 1st
- Grow received vectors
| Vectors | AmpR (μg/μL) | CmR (μg/μL) | KanR (μg/μL) |
|---|---|---|---|
| ECE112 | 100 | 0 | 0 |
| ECE147 | 50 | 0 | 0 |
| ECE149 | 50 | 0 | 0 |
| ECE150 | 50 | 0 | 0 |
| ECE151 | 50 | 0 | 0 |
| ECE153 | 100 | 0 | 0 |
| ECE162 | 100 | 0 | 0 |
| ECE165 | 100 | 10 | 0 |
| ECE166 | 100 | 10 | 0 |
| ECE171 | 50 | 0 | 10 |
| ECE172 | 100 | 10 | 0 |
| ECE176 | 100 | 5 | 0 |
- Preparation of Agar plates
We have bottles of 200mL.
| Type | Amp (μL) | Cm (μL) | Kan (μL) |
|---|---|---|---|
| Amp100 | 200 | 0 | 0 |
| Amp50 | 100 | 0 | 0 |
| Amp100 + Cm10 | 200 | 57.1 | 0 |
| Amp100 + Cm5 | 200 | 28.6 | 0 |
| Amp50 + Kan10 | 50 | 0 | 80 |
- Preparation of LB tubes
We prepare 10mL of LB in which we add a single colony.
| Type | Amp (μL) | Cm (μL) | Kan (μL) |
|---|---|---|---|
| Amp100 | 10 | 0 | 0 |
| Amp50 | 5 | 0 | 0 |
| Amp100 + Cm10 | 10 | 2.9 | 0 |
| Amp100 + Cm5 | 10 | 1.4 | 0 |
| Amp50 + Kan10 | 5 | 0 | 4 |
August 2nd
Yesterday's Results
- All Plates grew!
- ECE 172 seemed to have trouble growing on the plate (no single colonies, just one big lump at the start of the streak)
- ECE 150 grew too well! (possibly hard to pick out single colony)
- Tubes:
- ECE 176, 172, 153 seem to be quite clear!!
- ECE 150, 151 had strange floating 'colonies' in the tube
- All other tubes were quite cloudy
Lab Work
Fridged all plates except ECE 172 ==> Benched.
All tubes palleted and placed in freezer although ECE 176, 172, 153 doesn't seem to have any pallets
All red plates does not seem to have any extra growth despite the air conditioning had been turned off when I entered the lab this afternoon.
August 4th
Wet Work
- Check vectors
- for each vector, plasmid miniprep from frozen pellets (step 7 only once, with 60μL of Elution Buffer for ECE112, ECE147, ECE149, ECE 150 and only 30μL for the others)
- Single digest (15μL of SDW, 2μL of Buffer, 2μL of DNA, 1μL of enzyme, then incubate 10min at 37°C and 30 min for the one using HindIII with EcoRI buffer)
- Double digest (mix 7.5μL of each single digest, and then incubate 10min (or 30min for EcoRI + HindIII and EcoRI buffer) at 37°C, then heat shock 5min at 80°C
| Vectors | Enzyme 1 | Enzyme 2 | Buffer | Entire size | Size of part1 | Size of part2 |
|---|---|---|---|---|---|---|
| ECE112 | XbaI | EcoRI | Fast Digest | 10156 | 6900 | 3295 |
| ECE147 | EcoRI | HindIII | EcoRI | 5482 | 4913 | 600 |
| ECE149 | SpeI | PstI | Fast Digest | 6192 | 4919 | 1300 |
| ECE150 | SpeI | HimdIII | Buffer 2 | 6166 | 5300 | 778 |
| ECE151 | SpeI | EcoRI | Fast Digest | 6166 | 4825 | 1300 |
| ECE153 | SalI | Buffer 3 | ||||
| ECE162 | SalI | Buffer 3 | 7600 | 6000 | 1600 | |
| ECE165 | EcoRI | HindIII | EcoRI | 5952 | 5200 | 793 |
| ECE166 | EcoRI | HindIII | EcoRI | 7262 | 5100 | 2103 |
| ECE171 | EcoRI | PstI | Fast Digest | 5444 | 3600 | 1826 |
| ECE172 | HindIII | Buffer 2 | 6462 | 3900 | 2540 | |
| ECE176 | EcoRI | XbaI | Fast Digest | 866 | 4250 | 5128 |
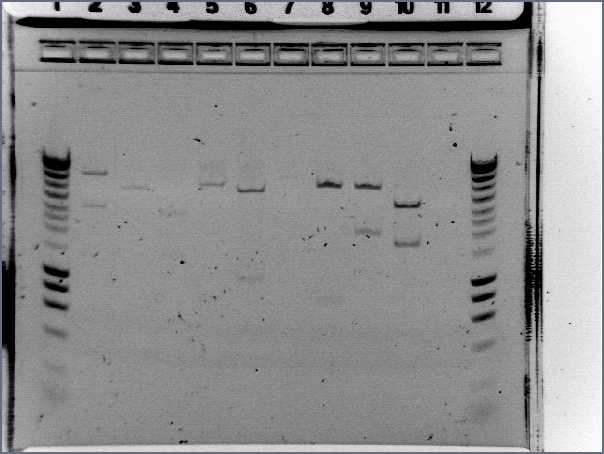
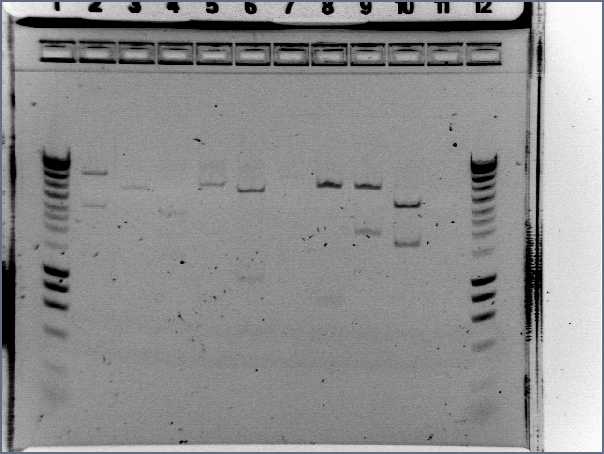
- Results
- Lane1 : HyperladderI
- Lane2 : ECE112
- Lane3 : ECE147
- Lane4 : ECE149
- Lane5 : ECE150
- Lane6 : ECE151
- Lane7 : ECE162
- Lane8 : ECE165
- Lane9 : ECE166
- Lane10 : ECE171
- Lane11 : ECE172
- Lane12 : HyperladderI
- ECE112, ECE165, ECE166, ECE171 : right bands, ok
- ECE172 : no band, not enough DNA
- ECE147, ECE150, ECE162 : only one band, maybe not enough DNA
- ECE149 : wrong size!
- ECE151 : not sure, check again
- Plates
- Streak new plates with different strains of Bacillus : 1A1, IA751 and IA771
August 5th
Checking vectors
- Plasmid miniprep for ECE166 (for stock), ECE172 and ECE153
- Digest of ECE153, ECE172 and ECE162 (with plasmid miniprep from yesterday)
- Run on a gel single digest for ECE147 (from yesterday with more DNA), ECE149 (from yesterday with more DNA), ECE150 (from yesterday with more DNA) and ECE 153, ECE162, ECE172
- Results
- Lane3 : HyperladderI
- Lane4 : ECE147
- Lane5 : ECE149
- Lane6 : ECE150
- Lane7 : ECE151
- Lane8 : ECE1162
- Lane9 : ECE172
- Lane10 : hyperladderI
Very low bands : not enough DNA!!!
Transformation of Bacillus
- Result of Nanodrop
| Vector | 260/280 | ng/μL |
|---|---|---|
| ECE112 | 1.75 | 64.6 |
| ECE166 | 172 | 138.6 |
- Prepare medium A with tryptophan, and medium B
- add 5mL of medium A in 3 different tubes, inoculate each tube with colonies (1A1, IA751 ans IA771)
- Check OD every 20min
- Incubate 90min
- Add 0.45mL of medium B and 0.05mL of culture in Ependorf tubes
- Incubate 90min
- Transform : add 1μg of DNA (some with ECE112, some with ECE166) (so you need to nanodrop samples before!)
- Incubate 30min
- Pipette 200μL of solution, spread it on aech plate, wait 10min, and do it again
- Incubate 24hours
- A few glycerol tubes to stock cells : add 2/7 glycerol to cell tubes
- Transformation from glycerol stock from 30/07/2008
- Spin glycerol stocks, pipette out glycerol
- Add 0.5mL of medium B, incubate for 1 hour
- Add 10μL of ECE112 (640ng)
- Incubate 2hours
- Plate 200μL, and 10min later, still 200μL
New stocks
- Do glycerol stock of I746001 and I746101 (no sterile glycerol)
- Put IA751, IA771 in 10mL LB
- Put ECE 176 in 10mL LB + antibiotic
- Reinoculate the tube of LB from yesterday with ECE166 plate
- ECE176 replated onto Amp100 + Cm5
August 6th
PCR parts of Agr operon
We received primers for Agr A,B,C, and D today - http://openwetware.org/wiki/IGEM:Cambridge/2008/Turing_Pattern_Formation/Primers
We must PCR each individual part out of the previous BioBricks (I746001 &I746101) in order to put our bacillus RBS on them.
I was concerned that the region of most homology would be the BioBrick prefix and suffix itself, so I first digested the DNA with Xbal and SpeI.
For some reason the digestion didn't work that well, so I've decided to go ahead and use the uncut BioBrick DNA as template for my reactions.
August 9th
Plans for Monday
Lab Work
- Be sure that the Spc50 plates are working, as the control for Spc50 grew and shouldn't have. We already know that things can die at Spc100 (the double crossover test from earlier this week) so it is unlikely that the stock is bad. We should test Spc50 again with regular bacillus and with E. coli at 50-100 ug/ml and hope that they die.
- Finish all the agr PCRs, but especially the RBS.
- Confirm that amyE insertion works, using the amylase/ery tests.
- Confirm that with ECE153 inserted, we get detectable fluoresence in a microscope.
- Confirm that ECE166 transformation works, using plasmid miniprepping.
- Find the qiagen miniprep kit or adapt the protocol.
- Confirm that when ECE166 is transformed, we get detectable fluoresence in a microscope.
Book/Computer Work
- Find good promoters in vectors and order PCR primers to get them out
- Find out how to assay for/make AIP
- Find/Contact Jim Ajioka
- Find out how to do a c-terminal GFP fusion for membrane proteins agrB and agrC
- Can we fuse GFP and still expect them to localize to the membrane?
- AgrB transmembrane topology (http://www.jbc.org/cgi/content/full/277/38/34736#F1). The signal peptide is not on the N terminus, and both the N and C terminus are intracellular.
- AgrC transmembrane topology (http://openurl.ingenta.com/content?genre=article&issn=0950-382X&volume=28&issue=3&spage=655&epage=662 (see figure 2 B)), the N terminus is probably extracellular. The C terminus is probably intracellular. The article doesn't say where the signal peptide is.
- Should we do a antibody tag instead?
- Can we fuse GFP and still expect them to localize to the membrane?
Longer Term Plans
- Ligate the RBS into a BB vector so we can build a RBS/GFP construct.
- Put the RBS/GFP construct into ECE166 and ECE 153.
- Compare the 'out of the box' ECE153 with ECE153 with RBS/GFP biobrick inserts
- Compare the 'out of the box' ECE166 with ECE166 with RBS/GFP biobrick inserts
August 11th
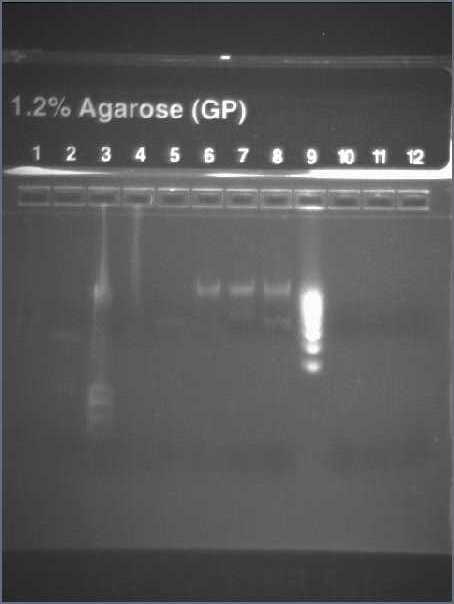
PCR Bacillus RBS
We created 2 different Bacillus RBS sites simply by primer annealing and extension. Each product includes an 8bp RBS plus the BioBrick prefix and suffix for a total of 54bp. The products were visualized on a 3.5% Agarose gel with bioline's hyperladder V.
RBSs is the consensus RBS sequence for Bacillus subtilis, and thus we believe it to be a very strong binding site - AAAGGAGG
RBSw has a 2bp modification from RBSs, which we believe will weaken it - AGAGGTGG
The amplification of RBSs yielded only one band on the gel. The product was purified using microclean and contained 14.8ng/uL after clean-up.
RBSw, however produced 2 bands on the gel. The correct size was gel-extracted and purified. After purification the yield was 12.3 ng/uL.
August 13th
PCR Agr A, B, C, D
Doing a PCR directly from the old Biobricks has worked for all parts of the operon. No prior digestion seems to be needed.
| Part | 260/280 | μg/mL |
|---|---|---|
| A | 1.78 | 92.5 |
| B | 1.87 | 96.5 |
| C | 1.9 | 127.7 |
| D | 1.82 | 26.3 |
August 15th
PCRing out promoters
- 4 different promoters : Pxyl, Pspc, Ppac, Pupp
- Make master mix enough for 4RXNs
Digest and Ligate Agr A and C to pSB4C5
Previous PCR products of Agr A and C were digested using Fermentas Fast Digest - EcoRI and PstI according to their protocol except a longer time was given to digest.
3uL of AgrA was used and 4.4 uL of AgrC was used and incubated for 40minutes.
10uL totaling to 1ug of pSB4C5 plasmid was also digested and incubated for 10 minutes.
The digestion was visualized on a 0.8% Agarose gel and produced expected sizes:
Plasmid: approx 3bk
Agr A: approx 700bp
Agr C: approx 1300bp
August 16th
Agr A and C ligation to pSB4C5
Ligation was performed using Fermentas Rapid Ligation kit according to their protocol.
After ligation, samples were visualized on a gel, but little product of the correct size was seen.
For agrA we should have gotten a band of size 3700 (3000plasmid + 700 insert)
For agrC we should have gotten a band of size 4300 (3000plasmid + 1300insert)
The bands of appropriate sizes were extracted for transformation.
August 18th
Check promoters
On Friday, PCR out promoters, we want to check them.
- Run a gel
- Results : good size of bands!!!
Transformation of AgrA and AgrC
AgrA and C gell-extracted ligation was transformed into Top 10 cells using standard protocol. Puc9 was used as a positive control.
|}
 "
"