Team:Harvard/Shewie
From 2008.igem.org
(→Molecular Biology with Shewanella oneidensis) |
(→Molecular Biology with Shewanella oneidensis) |
||
| Line 5: | Line 5: | ||
==Molecular Biology with <i>Shewanella oneidensis</i>== | ==Molecular Biology with <i>Shewanella oneidensis</i>== | ||
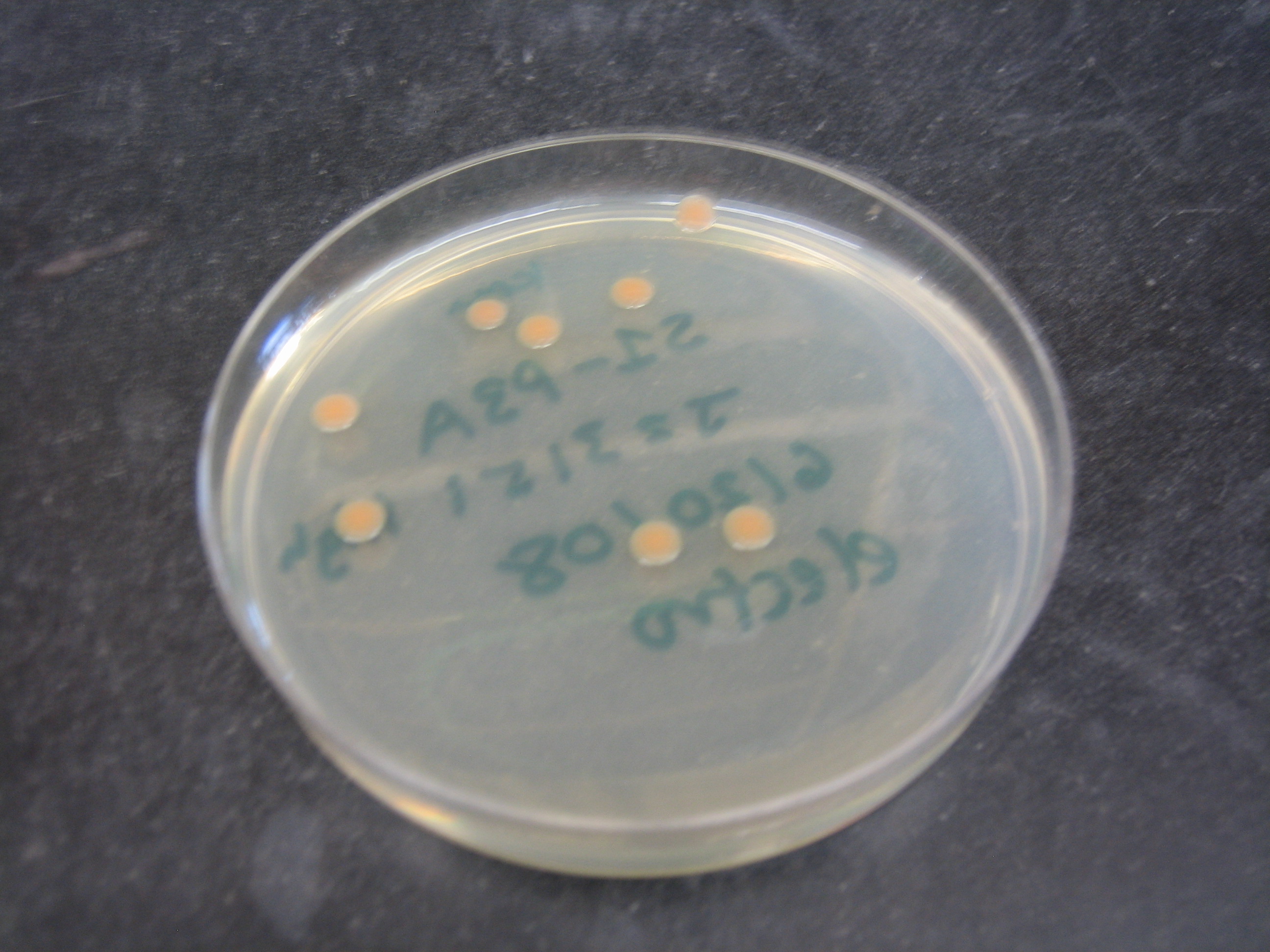
| - | [[Image:Shewanella colonies growing on plate.JPG|thumb| | + | [[Image:Shewanella colonies growing on plate.JPG|thumb|300px|''S. oneidensis MR-1'' colonies from a transformation]]The ''Shewanella oneidensis MR-1'' genome was sequenced in 2002, greatly increasing its usefulness as a model organism. It was found that it had a 4,969,803 base pair circular chromosome and a 161,613 base pair plasmid (Heidelberg et al. 2002). When cloning in ''S. oneidensis MR-1'', it has also been shown that plasmids with p15A origins replicate freely, whereas plasmids with a pMB1 origin of replication do not (Myers and Myers 1997). [We further found that …]. ''S. oneidensis MR-1'' grows at 30 ºC, can be electroporated (see protocol in our [[Team:Harvard/Notebook| Notebook]]) and forms round orange pink colonies on plates. It is resistant to ampicillin, but other resistance markers work (Daad). Together, these characteristics make ''S. oneidensis MR-1'' a genetically tractable organism good for exploring the possibilities of regulated bacterial electrical output. |
| + | |||
==Chassis Specifications== | ==Chassis Specifications== | ||
To facilitate standardization of different chassis, it may be advantageous to standardize a specification sheet. We provide a quick summary of ''S. oneidensis MR-1'' following what we think may be a suitable format: | To facilitate standardization of different chassis, it may be advantageous to standardize a specification sheet. We provide a quick summary of ''S. oneidensis MR-1'' following what we think may be a suitable format: | ||
Revision as of 19:05, 28 October 2008
Shewanella oneidensis MR-1
Shewanella oneidensis MR-1 is a gram-negative facultative anaerobe (Myers and Myers 1997). Under anaerobic conditions, it reduces a number of electron acceptors such as MN(IV). This ability can be harnessed by microbial fuel cells (MFC) to produce an electric current (Bretschger et al. 2007). When the bacteria are grown anaerobically in the anode chamber of an MFC, they release electrons onto the electrode, creating an electrical current. These diverse respiratory capabilities require a complex electron transport systems, including 39 c-type cytochromes (Heidelberg et al. 2002). These interesting characteristics of S. oneidensis MR-1 make it an important model organism for both studies of bioremediation as well as biotechnology applications.
Molecular Biology with Shewanella oneidensis
The Shewanella oneidensis MR-1 genome was sequenced in 2002, greatly increasing its usefulness as a model organism. It was found that it had a 4,969,803 base pair circular chromosome and a 161,613 base pair plasmid (Heidelberg et al. 2002). When cloning in S. oneidensis MR-1, it has also been shown that plasmids with p15A origins replicate freely, whereas plasmids with a pMB1 origin of replication do not (Myers and Myers 1997). [We further found that …]. S. oneidensis MR-1 grows at 30 ºC, can be electroporated (see protocol in our Notebook) and forms round orange pink colonies on plates. It is resistant to ampicillin, but other resistance markers work (Daad). Together, these characteristics make S. oneidensis MR-1 a genetically tractable organism good for exploring the possibilities of regulated bacterial electrical output.Chassis Specifications
To facilitate standardization of different chassis, it may be advantageous to standardize a specification sheet. We provide a quick summary of S. oneidensis MR-1 following what we think may be a suitable format:
 "
"