Imperial College/2 September 2008
From 2008.igem.org
(Difference between revisions)
m (New page: {{Imperial/StartPage}}__NOTOC__ {| cellpadding="10" border="0" |- valign="top" |{{#calendar: title=Imperial_College |year=2008 | month=07}} |{{#calendar: title=Imperial_College |year=2008 ...) |
|||
| (8 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{| cellpadding="10" border="0" | {| cellpadding="10" border="0" | ||
|- valign="top" | |- valign="top" | ||
| - | |||
|{{#calendar: title=Imperial_College |year=2008 | month=08}} | |{{#calendar: title=Imperial_College |year=2008 | month=08}} | ||
|{{#calendar: title=Imperial_College |year=2008 | month=09}} | |{{#calendar: title=Imperial_College |year=2008 | month=09}} | ||
| - | | rowspan="2" bgcolor=# | + | |{{#calendar: title=Imperial_College |year=2008 | month=10}} |
| + | | rowspan="2" bgcolor=#ffffff width="100%" | | ||
|} | |} | ||
| + | =2 September 2008= | ||
==Wet Lab== | ==Wet Lab== | ||
| + | ===Cloning=== | ||
| + | |||
| + | *Pfu PCR reactions for both AmyE integration sequences and the Laci gene were set up according to the standard [http://openwetware.org/index.php?title=IGEM:IMPERIAL/2008/Prototype/Wetlab/PCR protocol], using the optimal conditions determined yesterday and results run on a gel along with Aad9 Taq polymerase condiitons test (54<sup>o</sup>C, 52<sup>o</sup>C or 50<sup>o</sup>C for the first 10 cycles then 60<sup>o</sup>C for the last 20 cycles). Apropraite negativbe controls were used. For results, see tomorrow's entry. | ||
| + | *XL1-Blue cells containing the constructs in plasmids from GeneArt were grown up and incoulated into an overnight culture for midiprep tomorrow | ||
| + | |||
===Transformation of ''B.subtilis''=== | ===Transformation of ''B.subtilis''=== | ||
*In order to confirm the transformations of ''B.subtilis'' that were performed in the previous weeks, we have designed validation primers. These validation primers are complementary to regions flanking the amyE locus and as a results the PCR product size will reflect any inserted DNA within this region. Using these primers a single colony PCR was performed, (link to prorotcol for single colony PCR for verficication. | *In order to confirm the transformations of ''B.subtilis'' that were performed in the previous weeks, we have designed validation primers. These validation primers are complementary to regions flanking the amyE locus and as a results the PCR product size will reflect any inserted DNA within this region. Using these primers a single colony PCR was performed, (link to prorotcol for single colony PCR for verficication. | ||
| Line 32: | Line 38: | ||
**15-47 <sup>o</sup>C - Negative control - no DNA!!! | **15-47 <sup>o</sup>C - Negative control - no DNA!!! | ||
**16-51 <sup>o</sup>C - AmyE 1 positive control as previously shown to work. | **16-51 <sup>o</sup>C - AmyE 1 positive control as previously shown to work. | ||
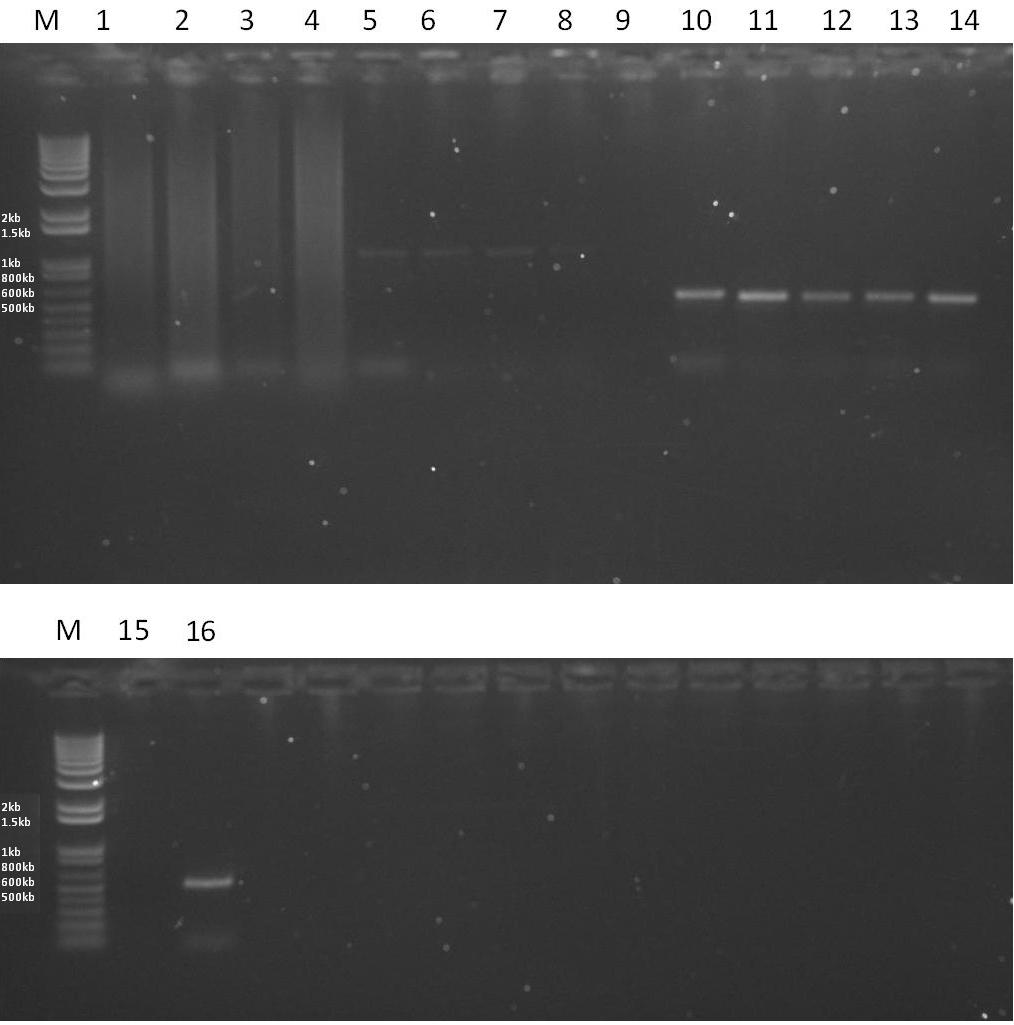
| + | *'''Results''' - From the gel below it can be seen that the EpsE PCR reactions have worked, however with slight levels of contamination when a high annealing temperature is used. The XylR PCR reaction has not work efficiently, clearly visible from the faint bands. This means that the temperatures for annealing need to be optimised further. | ||
| + | |||
| + | ===Results=== | ||
| + | [[Image:RESULTS2.JPG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer.]] | ||
| + | <br> | ||
| + | {{Imperial/EndPage|Notebook|Notebook}} | ||
Latest revision as of 20:43, 28 October 2008
2 September 2008Wet LabCloning
Transformation of B.subtilis
Testing of Primers
Results
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"