Imperial College/2 September 2008
From 2008.igem.org
(Difference between revisions)
m (→Wet Lab) |
|||
| (One intermediate revision not shown) | |||
| Line 12: | Line 12: | ||
===Cloning=== | ===Cloning=== | ||
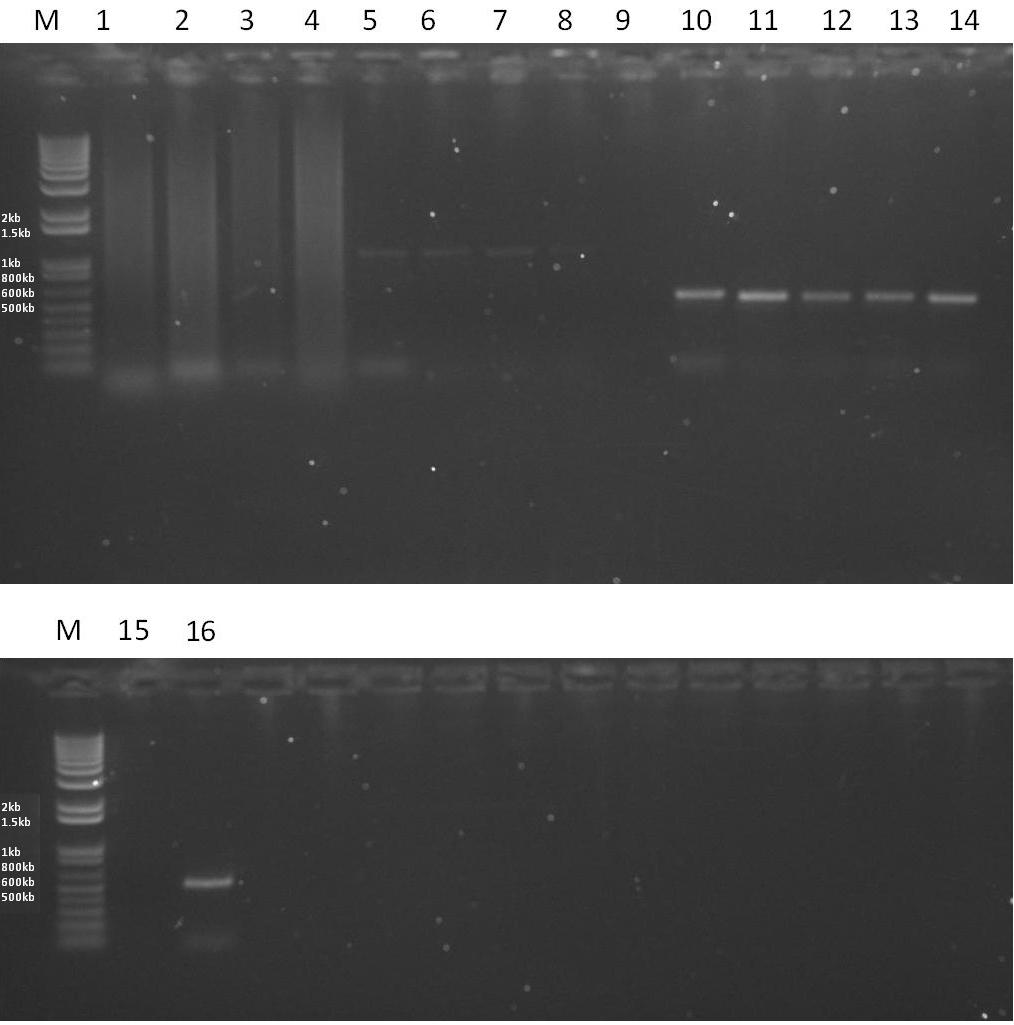
| - | *Pfu PCR reactions for both AmyE integration sequences and the Laci gene were set up according to the standard | + | *Pfu PCR reactions for both AmyE integration sequences and the Laci gene were set up according to the standard [http://openwetware.org/index.php?title=IGEM:IMPERIAL/2008/Prototype/Wetlab/PCR protocol], using the optimal conditions determined yesterday and results run on a gel along with Aad9 Taq polymerase condiitons test (54<sup>o</sup>C, 52<sup>o</sup>C or 50<sup>o</sup>C for the first 10 cycles then 60<sup>o</sup>C for the last 20 cycles). Apropraite negativbe controls were used. For results, see tomorrow's entry. |
*XL1-Blue cells containing the constructs in plasmids from GeneArt were grown up and incoulated into an overnight culture for midiprep tomorrow | *XL1-Blue cells containing the constructs in plasmids from GeneArt were grown up and incoulated into an overnight culture for midiprep tomorrow | ||
| Line 42: | Line 42: | ||
===Results=== | ===Results=== | ||
[[Image:RESULTS2.JPG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer.]] | [[Image:RESULTS2.JPG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer.]] | ||
| + | <br> | ||
| + | {{Imperial/EndPage|Notebook|Notebook}} | ||
Latest revision as of 20:43, 28 October 2008
2 September 2008Wet LabCloning
Transformation of B.subtilis
Testing of Primers
Results
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"