|
|
| (7 intermediate revisions not shown) |
| Line 9: |
Line 9: |
| | | | |
| | =5 September 2008= | | =5 September 2008= |
| - | ==Wetlab== | + | =='''Wetlab'''== |
| | | | |
| | ===Cloning=== | | ===Cloning=== |
| Line 23: |
Line 23: |
| | | | |
| | ===Trial Run: ''B. subtilis'' Growth Curve=== | | ===Trial Run: ''B. subtilis'' Growth Curve=== |
| - | In our second trial run to obtain growth and calibration curves for ''B. subtilis'', we cultured our "B. subtilis" for two days and continously monitored the changes in OD600. By plotting OD600 against Time/h, we obtained the following growth curve for "B. subtilis". | + | In our second trial run to obtain growth and calibration curves for ''B. subtilis'', we cultured our "B. subtilis" for two days and monitored the changes in OD<sub>600</sub> continuously. By plotting OD<sub>600</sub> against time/h, we obtained the following growth curve for "B. subtilis". |
| | | | |
| | [[Image:Trial_Run_Growth_Curve.png|thumb|600px|center|Growth Curve for ''B. subtilis'' from trial run]] | | [[Image:Trial_Run_Growth_Curve.png|thumb|600px|center|Growth Curve for ''B. subtilis'' from trial run]] |
| Line 29: |
Line 29: |
| | ===Calibration Curve=== | | ===Calibration Curve=== |
| | | | |
| - | For six of the data points from the above growth curve, we plated 0.1 ml of ''B. subtilis'' on LB agar plates and performed serial dilution in order to calculate the colony-forming units (CFU per ml) for the corresponding OD<sub>600</sub>. However, we only managed to obtain two sets of valid data. The ''B. subtilis'' mixtures we plated for higher values of OD600 were too dilute, hence no bacterial growth was observed on these LB agar plates. | + | For six of the data points from the above growth curve, we plated 0.1 ml of ''B. subtilis'' on LB agar plates and performed serial dilutions in order to calculate the colony-forming units (CFU per ml) for the corresponding OD<sub>600</sub>. However, we only managed to obtain two sets of valid data. The ''B. subtilis'' mixtures we plated for higher values of OD600 were too dilute, hence no bacterial growth was observed on these LB agar plates. |
| | | | |
| | [[Image:Trial_Run_B_Subtilis_Calibration_Curve.png|thumb|600px|center|Growth Curve for ''B. subtilis'' from trial run]] | | [[Image:Trial_Run_B_Subtilis_Calibration_Curve.png|thumb|600px|center|Growth Curve for ''B. subtilis'' from trial run]] |
| Line 43: |
Line 43: |
| | *Check OD<sub>600</sub> readings more frequently during the exponential growth phase. This would ensure a more even distribution of data points for the calibration curve. | | *Check OD<sub>600</sub> readings more frequently during the exponential growth phase. This would ensure a more even distribution of data points for the calibration curve. |
| | | | |
| - | ==Dry Lab== | + | =='''Dry Lab'''== |
| | | | |
| | ===Motility=== | | ===Motility=== |
| - | [[Image:RunTumble.jpg|300px|thumb|center|Complex Plot of Velocity and Angle]]
| |
| | *10 cells were tracked manually from Video 3. The graph of Z=Rexp(i*X) was plotted, R being the displacement and X being the angle between three consecutive coordinate points. It is hypothesised that tumbling will occur with very low displacement and high angle, while running occurs with large displacement and low angle. | | *10 cells were tracked manually from Video 3. The graph of Z=Rexp(i*X) was plotted, R being the displacement and X being the angle between three consecutive coordinate points. It is hypothesised that tumbling will occur with very low displacement and high angle, while running occurs with large displacement and low angle. |
| | *It was found that segmentation could not be done visually. However, using subtractive clustering analysis in the "Find Cluster Toolbox" in MATLAB, we were able to identify 2 centers of clustering, representing tumbling and running. However, more data has to be obtained to visually identify running and tumbling clusters. | | *It was found that segmentation could not be done visually. However, using subtractive clustering analysis in the "Find Cluster Toolbox" in MATLAB, we were able to identify 2 centers of clustering, representing tumbling and running. However, more data has to be obtained to visually identify running and tumbling clusters. |
| | + | [[Image:Cluster.jpg|600px|thumb|center|Complex Plot of Velocity*exp(i*Angle)]] |
| | | | |
| | ===Microscopy=== | | ===Microscopy=== |
| | 2 microscopy sessions will be arranged for next week, on Wednesday and Friday. | | 2 microscopy sessions will be arranged for next week, on Wednesday and Friday. |
| | | | |
| - | ===Genetic Circuit===
| |
| | | | |
| - | ===Growth===
| + | <br> |
| - | | + | {{Imperial/EndPage|Notebook|Notebook}} |
| - | {{Imperial/EndPage}} | + | |
|
| August
|
| M | T | W | T | F | S | S
|
|
|
|
|
| [http://2008.igem.org/Imperial_College/1_August_2008 1]
| [http://2008.igem.org/Imperial_College/2_August_2008 2]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/3_August_2008&action=edit 3]
|
| [http://2008.igem.org/Imperial_College/4_August_2008 4]
| [http://2008.igem.org/Imperial_College/5_August_2008 5]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/6_August_2008&action=edit 6]
| [http://2008.igem.org/Imperial_College/7_August_2008 7]
| [http://2008.igem.org/Imperial_College/8_August_2008 8]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/9_August_2008&action=edit 9]
| [http://2008.igem.org/Imperial_College/10_August_2008 10]
|
| [http://2008.igem.org/Imperial_College/11_August_2008 11]
| [http://2008.igem.org/Imperial_College/12_August_2008 12]
| [http://2008.igem.org/Imperial_College/13_August_2008 13]
| [http://2008.igem.org/Imperial_College/14_August_2008 14]
| [http://2008.igem.org/Imperial_College/15_August_2008 15]
| [http://2008.igem.org/Imperial_College/16_August_2008 16]
| [http://2008.igem.org/Imperial_College/17_August_2008 17]
|
| [http://2008.igem.org/Imperial_College/18_August_2008 18]
| [http://2008.igem.org/Imperial_College/19_August_2008 19]
| [http://2008.igem.org/Imperial_College/20_August_2008 20]
| [http://2008.igem.org/Imperial_College/21_August_2008 21]
| [http://2008.igem.org/Imperial_College/22_August_2008 22]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/23_August_2008&action=edit 23]
| [http://2008.igem.org/Imperial_College/24_August_2008 24]
|
| [http://2008.igem.org/Imperial_College/25_August_2008 25]
| [http://2008.igem.org/Imperial_College/26_August_2008 26]
| [http://2008.igem.org/Imperial_College/27_August_2008 27]
| [http://2008.igem.org/Imperial_College/28_August_2008 28]
| [http://2008.igem.org/Imperial_College/29_August_2008 29]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/30_August_2008&action=edit 30]
| [http://2008.igem.org/Imperial_College/31_August_2008 31]
|
|
| September
|
| M | T | W | T | F | S | S
|
| [http://2008.igem.org/Imperial_College/1_September_2008 1]
| [http://2008.igem.org/Imperial_College/2_September_2008 2]
| [http://2008.igem.org/Imperial_College/3_September_2008 3]
| [http://2008.igem.org/Imperial_College/4_September_2008 4]
| [http://2008.igem.org/Imperial_College/5_September_2008 5]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/6_September_2008&action=edit 6]
| [http://2008.igem.org/Imperial_College/7_September_2008 7]
|
| [http://2008.igem.org/Imperial_College/8_September_2008 8]
| [http://2008.igem.org/Imperial_College/9_September_2008 9]
| [http://2008.igem.org/Imperial_College/10_September_2008 10]
| [http://2008.igem.org/Imperial_College/11_September_2008 11]
| [http://2008.igem.org/Imperial_College/12_September_2008 12]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/13_September_2008&action=edit 13]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/14_September_2008&action=edit 14]
|
| [http://2008.igem.org/Imperial_College/15_September_2008 15]
| [http://2008.igem.org/Imperial_College/16_September_2008 16]
| [http://2008.igem.org/Imperial_College/17_September_2008 17]
| [http://2008.igem.org/Imperial_College/18_September_2008 18]
| [http://2008.igem.org/Imperial_College/19_September_2008 19]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/20_September_2008&action=edit 20]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/21_September_2008&action=edit 21]
|
| [http://2008.igem.org/Imperial_College/22_September_2008 22]
| [http://2008.igem.org/Imperial_College/23_September_2008 23]
| [http://2008.igem.org/Imperial_College/24_September_2008 24]
| [http://2008.igem.org/Imperial_College/25_September_2008 25]
| [http://2008.igem.org/Imperial_College/26_September_2008 26]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/27_September_2008&action=edit 27]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/28_September_2008&action=edit 28]
|
| [http://2008.igem.org/Imperial_College/29_September_2008 29]
| [http://2008.igem.org/Imperial_College/30_September_2008 30]
|
|
| October
|
| M | T | W | T | F | S | S
|
|
|
| [http://2008.igem.org/Imperial_College/1_October_2008 1]
| [http://2008.igem.org/Imperial_College/2_October_2008 2]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/3_October_2008&action=edit 3]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/4_October_2008&action=edit 4]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/5_October_2008&action=edit 5]
|
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/6_October_2008&action=edit 6]
| [http://2008.igem.org/Imperial_College/7_October_2008 7]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/8_October_2008&action=edit 8]
| [http://2008.igem.org/Imperial_College/9_October_2008 9]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/10_October_2008&action=edit 10]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/11_October_2008&action=edit 11]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/12_October_2008&action=edit 12]
|
| [http://2008.igem.org/Imperial_College/13_October_2008 13]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/14_October_2008&action=edit 14]
| [http://2008.igem.org/Imperial_College/15_October_2008 15]
| [http://2008.igem.org/Imperial_College/16_October_2008 16]
| [http://2008.igem.org/Imperial_College/17_October_2008 17]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/18_October_2008&action=edit 18]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/19_October_2008&action=edit 19]
|
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/20_October_2008&action=edit 20]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/21_October_2008&action=edit 21]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/22_October_2008&action=edit 22]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/23_October_2008&action=edit 23]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/24_October_2008&action=edit 24]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/25_October_2008&action=edit 25]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/26_October_2008&action=edit 26]
|
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/27_October_2008&action=edit 27]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/28_October_2008&action=edit 28]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/29_October_2008&action=edit 29]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/30_October_2008&action=edit 30]
| [http://2008.igem.org/wiki/index.php?title=Imperial_College/31_October_2008&action=edit 31]
|
|
|
5 September 2008
Wetlab
Cloning
- PCR reaction to clone the LacI gene using Pfu set up - standard condtitions and reagents[http://openwetware.org/wiki/IGEM:IMPERIAL/2008/Prototype/Wetlab/PCR]; 56°C for the first 10 cycles, 65°C for the 20 cycles.
- Testing PCR reactions (3) for the Biobrick insert size verification primers (with the pSB1AK3 vector) set up - taq conditions and reagents; at 60°C, 58°C or 56°C for 30 cycles
- GeneArt construct 7, 8, 9 and 11 digests, CAT digest and pSB1A2 digest and the insert (excluding for pSB1A2)purified by gel-purification (pSB1A2 vector minus insert was purified)
- AmyE 5', AmyE 3' and Aad9 PCR reactions digested with XbaI and SpeI for cloning into Biobricks and PCR purified
- Terminator Biobrick digested and PCR purified to remove Prefix fragment
Colony Counting
After re-calibrating our illuminator, it can now count colonies with a fair degree of accuracy. Manual addition and removal of points allows us to fine-tune the results, so we will use this method from now on.
Trial Run: B. subtilis Growth Curve
In our second trial run to obtain growth and calibration curves for B. subtilis, we cultured our "B. subtilis" for two days and monitored the changes in OD600 continuously. By plotting OD600 against time/h, we obtained the following growth curve for "B. subtilis".
 Growth Curve for B. subtilis from trial run Calibration Curve
For six of the data points from the above growth curve, we plated 0.1 ml of B. subtilis on LB agar plates and performed serial dilutions in order to calculate the colony-forming units (CFU per ml) for the corresponding OD600. However, we only managed to obtain two sets of valid data. The B. subtilis mixtures we plated for higher values of OD600 were too dilute, hence no bacterial growth was observed on these LB agar plates.
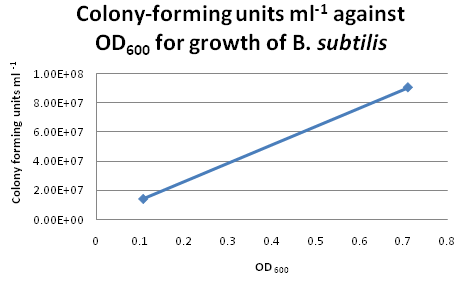 Growth Curve for B. subtilis from trial run Suggestions for Further Improvements
For the official experiment to obtain our calibration curve, we should take the following precautions:
- Pipette tips should be changed after each transfer of solution during serial dilution. This would avoid contamination and produce more accurate results for the calibration curve.
- Choose dilution factors for plating more carefully. It is not advisable to choose very high dilution factors e.g. those higher than 10E8.
- Check OD600 readings more frequently during the exponential growth phase. This would ensure a more even distribution of data points for the calibration curve.
Dry Lab
Motility
- 10 cells were tracked manually from Video 3. The graph of Z=Rexp(i*X) was plotted, R being the displacement and X being the angle between three consecutive coordinate points. It is hypothesised that tumbling will occur with very low displacement and high angle, while running occurs with large displacement and low angle.
- It was found that segmentation could not be done visually. However, using subtractive clustering analysis in the "Find Cluster Toolbox" in MATLAB, we were able to identify 2 centers of clustering, representing tumbling and running. However, more data has to be obtained to visually identify running and tumbling clusters.
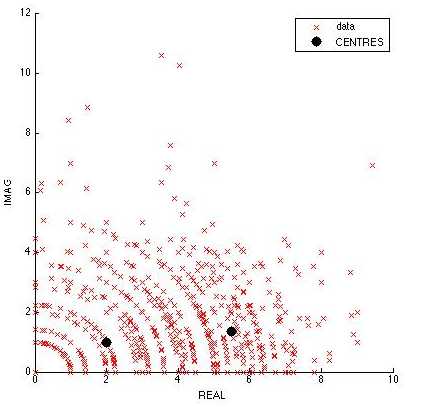 Complex Plot of Velocity*exp(i*Angle) Microscopy
2 microscopy sessions will be arranged for next week, on Wednesday and Friday.
|
 "
"



