Team:NTU-Singapore/Wetlab/Experimental Results
From 2008.igem.org
Lalala8585 (Talk | contribs) (New page: <html><link rel="stylesheet" href="http://greenbear88.googlepages.com/ntu_igem.css" type="text/css"></html> <div id="header">{{User:Greenbear/sandbox/header}}</div> <div id="mainconten...) |
Lalala8585 (Talk | contribs) |
||
| (11 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<div id="maincontent" style="margin-top:250px;"> | <div id="maincontent" style="margin-top:250px;"> | ||
| - | |||
| - | |||
| - | + | ='''Verification of Detection & Lysis system'''= | |
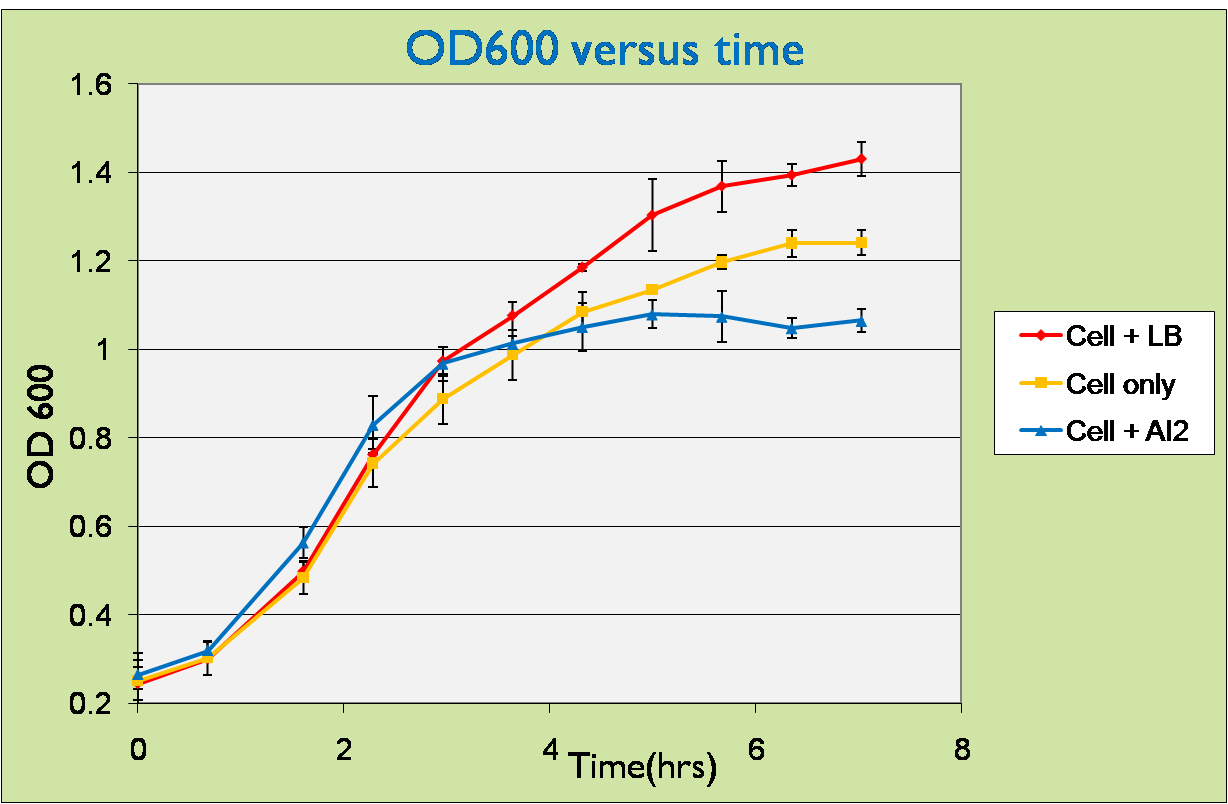
| + | As stated before, we wish to verify that the cells with the pLsrA-Lysis system would lyse upon Ai-2 inoculation. We used the OD of the cells as a form of verification in this experiment. | ||
| - | + | =='''OD600 measurement of pLsrA-Lysis-containing LuxS mutant upon addition of AI-2'''== | |
| - | + | ||
| - | + | ||
| - | [[Image: | + | [[Image:OD time.png|thumb|center|800px|Changing OD]]<br/> |
| - | + | For this experiment, samples of LuxS mutants carrying pLsrA-lysis plasmid were introduced with supernatants containing AI-2 and the optical density was measured. As pure AI-2 are very hard to obtain, we harvested them indirectly by taking the supernatants of the wildtype bacteria after centrifuged and filtering through 0.2µm pore size filters (refer to [https://2008.igem.org/Team:NTU-Singapore/Wetlab/Materials_and_Equipment Materials and Equipment] for more details). So the supernatants that we used for testing do not contain only AI-2 but also LB broth. Therefore, for all our testing experiments, we used bacteria added with LB as negative control. <br/> | |
| - | + | We can observe that initially, the curves of cell samples with LB and AI-2 have slightly higher slopes than that of sample with cells only. This can be easily understood because the supernatants also contain considerable amounts of LB, which promotes cell growth.<br/> | |
| + | However, after about 3 hours, the cell induced with AI-2 reached steady state while the control samples were still growing. This can be well explained by the effect of lysis, which counters cell growth at the same time. | ||
| + | Therefore, the cells in fact lyzed under the induction by AI-2. Or in other words, '''our detection system works as expected!!''' | ||
| + | <br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| - | == | + | var tags_before_clock = "<b>It is now " |
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| - | == | + | if(navigator.appName == "Netscape") { |
| - | + | document.write('<layer id="clock"></layer><br>'); | |
| - | + | } | |
| - | + | if (navigator.appVersion.indexOf("MSIE") != -1){ | |
| - | + | document.write('<span id="clock"></span>'); | |
| - | + | } | |
| - | + | ||
| - | + | ||
| - | + | DaysofWeek = new Array() | |
| - | + | DaysofWeek[0]="Sunday" | |
| - | [[ | + | DaysofWeek[1]="Monday" |
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| - | + | Months = new Array() | |
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | setInterval("upclock()",1000); | ||
| + | //--> | ||
| + | </script> | ||
| + | </html> | ||
Latest revision as of 23:54, 28 October 2008
|
Verification of Detection & Lysis system
As stated before, we wish to verify that the cells with the pLsrA-Lysis system would lyse upon Ai-2 inoculation. We used the OD of the cells as a form of verification in this experiment.
OD600 measurement of pLsrA-Lysis-containing LuxS mutant upon addition of AI-2
For this experiment, samples of LuxS mutants carrying pLsrA-lysis plasmid were introduced with supernatants containing AI-2 and the optical density was measured. As pure AI-2 are very hard to obtain, we harvested them indirectly by taking the supernatants of the wildtype bacteria after centrifuged and filtering through 0.2µm pore size filters (refer to Materials and Equipment for more details). So the supernatants that we used for testing do not contain only AI-2 but also LB broth. Therefore, for all our testing experiments, we used bacteria added with LB as negative control.
We can observe that initially, the curves of cell samples with LB and AI-2 have slightly higher slopes than that of sample with cells only. This can be easily understood because the supernatants also contain considerable amounts of LB, which promotes cell growth.
However, after about 3 hours, the cell induced with AI-2 reached steady state while the control samples were still growing. This can be well explained by the effect of lysis, which counters cell growth at the same time.
Therefore, the cells in fact lyzed under the induction by AI-2. Or in other words, our detection system works as expected!!
 "
"