Team:UC Berkeley Tools/Development
From 2008.igem.org
(→PoBoL: BioBrick Data Sharing Made Easy) |
|||
| Line 27: | Line 27: | ||
==PoBoL: BioBrick Data Sharing Made Easy== | ==PoBoL: BioBrick Data Sharing Made Easy== | ||
| - | [[Image:PoBoLDiagram.png|thumb|left|200 px|<span style="color: | + | [[Image:PoBoLDiagram.png|thumb|left|200 px|<span style="color: white"><small>''A diagram representing the basic structural concepts behind PoBoL''</small></span>]] |
| - | PoBoL stands for "Provisional BioBrick Language", and is the name given to a technical standard for BioBrick data in synthetic biology databases. Put simply, our implementation of PoBoL | + | PoBoL stands for "Provisional BioBrick Language", and is the name given to a technical standard for BioBrick data in synthetic biology databases. Put simply, our implementation of PoBoL is encapsulated in the Clotho Data structure. We indicate that this is our version since it adds objects which are not part of PoBoL. These include collections of BioBricks and composite information (right and left children and order). One of the big issues current facing synthetic biology is coming up with a parts standardization. We hope that Clotho can help to raise and solve issues in this area by being a deployment vehicle for ideas. Since we have created Clotho in a highly modular way, it can be the test bed for a variety of part standardization ideas. |
| - | + | ||
For more information on the PoBoL standard, check out [http://www.pobol.org/ www.pobol.org] | For more information on the PoBoL standard, check out [http://www.pobol.org/ www.pobol.org] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Increasing Collaboration Through Better Access To Database== | ==Increasing Collaboration Through Better Access To Database== | ||
Through a combination of PoBoL | Through a combination of PoBoL | ||
Revision as of 08:04, 29 October 2008
Clotho Beta: Future Development
The next release of Clotho after iGEM is planned for a late Spring 2009 release. This will be the official Beta release (the other releases will have been Alpha and iGEM). The main focus of the release will be to fully implement all the features currently in Clotho as well as fix bugs that are found. Clotho testing and deployment will begin with Anderson Lab at UCB and expand to Voigt Lab at UCSF. It is our goal to have Clotho be a main software component in each lab. In addition to general robustness, Clotho Beta should also incorporate a few new features as will be described in the following sections:
New Features
- New design views to supplement the Sequence View and allow easier designing of basic and composite BioBrick parts. This will likely include a "schematic view" for assembling parts, similar to the one in [http://web.mit.edu/jagoler/www/biojade/overview.html BioJade]. We would also like to explore a "parts view" where the individual parts have associated icons. A design now no longer becomes writing nucleotide sequences but rather dragging around icons which have sequence data associated with them.
- Ability to interface with devices, including robots that can be used for automated production of parts designed in Clotho. (In the future, this will contain a link to Anderson Lab's Biomek robot.) This aspect will be tied to the assembly algorithms already in Clotho. These algorithms will be expanded upon to deal with other assembly issues such as tracking methylated "righty" and "lefty" parts.
- Increased support for other interchange format standards such as [http://www.sequenceontology.org/gff3.shtml GFF3]. This will allow us to begin to interface with such tools as BioStudio.
Improved Features:
- More database support: we plan to expand the number and variety of databases that Clotho can connect to, including [http://brickit.wiki.sourceforge.net/ BrickIt!]) and the JBEI registry. One of the databases at the top of our priority list is iGEM's own [http://partsregistry.org/Main_Page Registry of Standard Biological Parts].
- Improved Plug-in environment that allows users to add even more expanded functionality to Clotho. This will include more extension points where plug-ins can be integrated and a set of more fully featured interfaces for existing extensions.
Helping Out In The Lab: Assembly Algorithms
- One of the initial algorithms we added to Clotho was an "optimal assembly" algorithm that can take in a list of goal composite parts and return a list with the minimum number of steps required to build those parts - a particularly useful feature if the goal parts utilize many of the same basic parts as subcomponents. The UC Berkeley wet team used these tools on a collection of 496 goal composite parts. Had no overlap been found (e.g. using naive methods), 5961 total parts would have to have been made. The program generated an assembly tree that required only 1283 total parts be built. That is a savings of 4678 parts. This assembly took just over 4 minutes to complete using the algorithm. If this same process were attempted by hand, it would take multiple people over 3 days to complete.
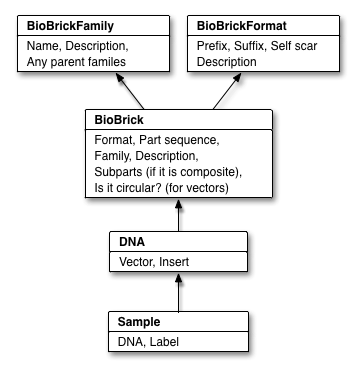
PoBoL: BioBrick Data Sharing Made Easy
PoBoL stands for "Provisional BioBrick Language", and is the name given to a technical standard for BioBrick data in synthetic biology databases. Put simply, our implementation of PoBoL is encapsulated in the Clotho Data structure. We indicate that this is our version since it adds objects which are not part of PoBoL. These include collections of BioBricks and composite information (right and left children and order). One of the big issues current facing synthetic biology is coming up with a parts standardization. We hope that Clotho can help to raise and solve issues in this area by being a deployment vehicle for ideas. Since we have created Clotho in a highly modular way, it can be the test bed for a variety of part standardization ideas.
For more information on the PoBoL standard, check out [http://www.pobol.org/ www.pobol.org]
Increasing Collaboration Through Better Access To Database
Through a combination of PoBoL
 "
"