Team:Newcastle University/Workbench
From 2008.igem.org
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Newcastle University/Header}} | {{:Team:Newcastle University/Header}} | ||
| - | {{:Team:Newcastle University/Template: | + | {{:Team:Newcastle University/Template:UnderTheSoftware |
|page-title=[[Team:Newcastle University/Workbench|Workbench]]}} | |page-title=[[Team:Newcastle University/Workbench|Workbench]]}} | ||
| + | <div id="maincol"> | ||
==Workbench== | ==Workbench== | ||
| - | + | <html><a name="aims"></html> | |
| - | ===Aims:=== | + | ===Aims and Objectives :=== |
The proposed Synthetic Biology Workbench (cool name needed) will incorporate the functionality of a biological network modeling and simulation program along with spiffy new algorithms that can be used in modern synthetic biology. This will enable users to finetune the expression of models suitable for their purposes, particularly for such fun activities like iGEM competitions. | The proposed Synthetic Biology Workbench (cool name needed) will incorporate the functionality of a biological network modeling and simulation program along with spiffy new algorithms that can be used in modern synthetic biology. This will enable users to finetune the expression of models suitable for their purposes, particularly for such fun activities like iGEM competitions. | ||
| Line 11: | Line 12: | ||
Once the biological circuit is ready in principle, it can be put into practice. The user will be able to request the optimal biological circuit, composed from BioBricks from the repository. | Once the biological circuit is ready in principle, it can be put into practice. The user will be able to request the optimal biological circuit, composed from BioBricks from the repository. | ||
| + | </div> | ||
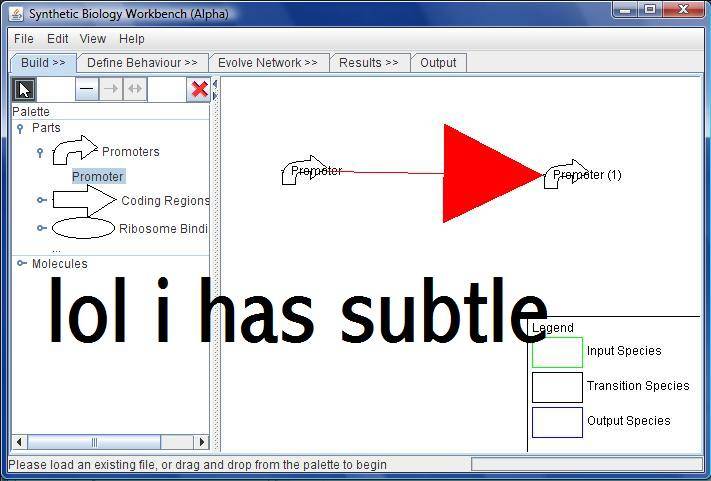
| + | [[Image:Subtle.jpg|thumb|right|DiS with a very subtle directed line, in a prototype version.]] | ||
| + | Our objectives are to: | ||
| + | * get a model | ||
| + | * load from SBML and/or CellML | ||
| + | * create using [[Team:Newcastle University/Parts Repository|parts]] and [[Team:Newcastle University/Constraints Repository|constraints]] | ||
| + | * show the model | ||
| + | * network view | ||
| + | * basic parts view. Matt suggested being able to view both strands and flip them around. | ||
| + | * take in conditions for synthetic evolution and send models to the [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] | ||
| + | * output the sequence in a standard format | ||
| - | === | + | <html><a name="user-stories"></html> |
| - | + | ===User Stories=== | |
| - | + | ====Parts Repository==== | |
| - | + | # The program must display a list of all available parts. The program requests a hierarchical list of all part names, part types, and their part ids from the Parts Repository (PR). Upon receiving the list, the program sorts the parts into different categories based on the part type, chooses a display icon for each type, and presents the list of part names with their part type icons to the user. | |
| - | + | # The user wishes to change the type of chassis. The program retrieves a list of all chassis names and chassis ids from the PR and presents it to the user. | |
| - | + | # The program must display a list of all types of parts. It requests a list of all part types and their part type ids from the PR. Then it displays the list of part names to the user. | |
| - | + | # The program must display all of the information about a single part. It gives the part id to the PR and receives a full description of the part in return. It then displays the part information to the user. | |
| - | + | # The program must save a CellML model of a part. It gives the part id to the PR and receives a segment of CellML in return. It then saves the CellML to file. | |
| - | + | ||
| + | ====Constraints Repository==== | ||
| + | # The program must show all possible interactions to a particular part. It passes the part id to the Constraints Repository and receives a list of all possible interaction ids in return. It then retrieves the details of each interaction with the interaction id. It then shows the possible interactions to the user. | ||
| + | ====Evolutionary Algorithm==== | ||
| + | # The user finally wants to evolve her model in silico. The program collects the starting model, composed of a series of part ids, and the proposed behaviour in the form of a truth table, and gives it to the Evolutionary Algorithm (EA) for evolution. The program shows a “busy working” screen while the Evolutionary Algorithm computes the best model. When the EA finishes, the program receives the proposed best model(s) and displays them, along with their behaviour, to the user. | ||
| + | |||
| + | ---- | ||
| + | <html><a name="outcomes"></html> | ||
| + | ==Outcome== | ||
| + | |||
| + | [[Image:Wb-layout.jpg|300px|thumb|The architecture of DiS is based on certain software engineering principles. Certain design patterns, such as Singleton, have been used extensively]] | ||
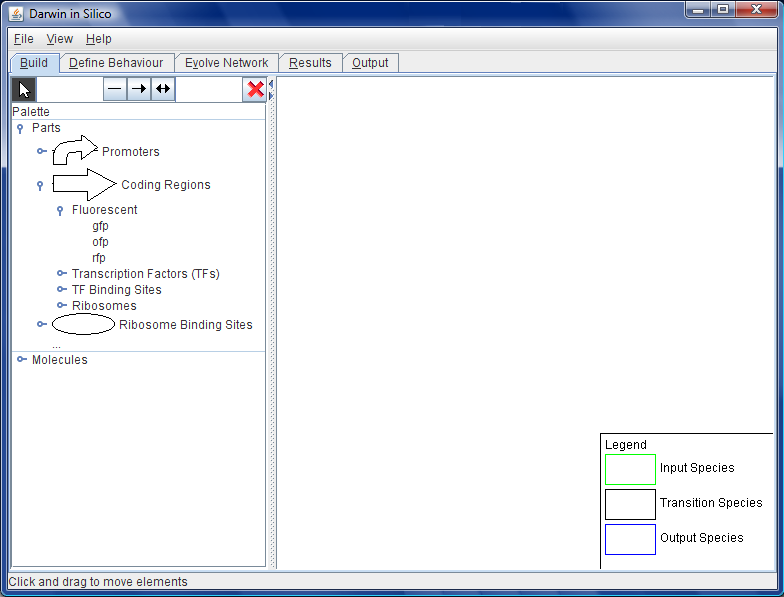
| + | [[Image:Wb.png|thumb|200px|right|Figure A: The opening screen of Darwin in Silico - the Synthetic Biology Workbench. Here, we see the first part of the workflow, the ''Build'', where the initial structure of the proposed circuit is laid out.]] | ||
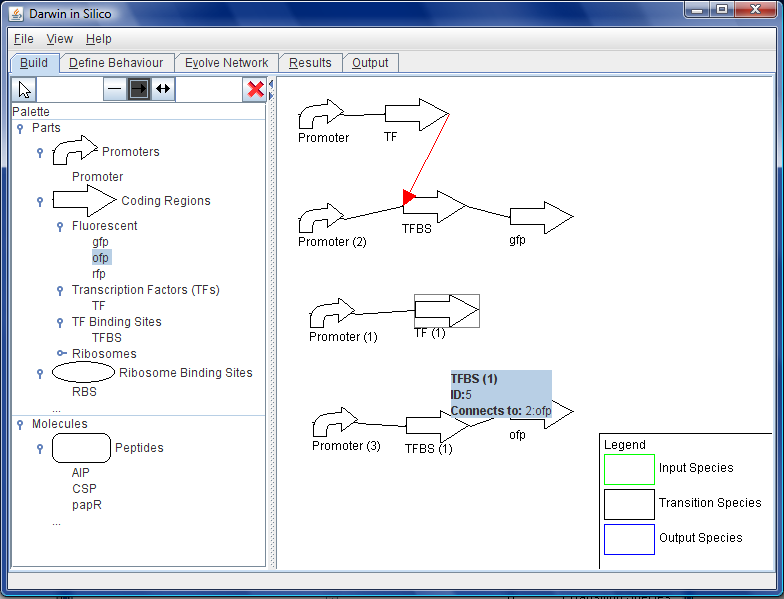
| + | [[Image:Wb-connections.png|200px|thumb|right|Figure B: Parts can be dragged from the left pane onto the building area, and connected with different kinds of arrows to indicate different relationships.]] | ||
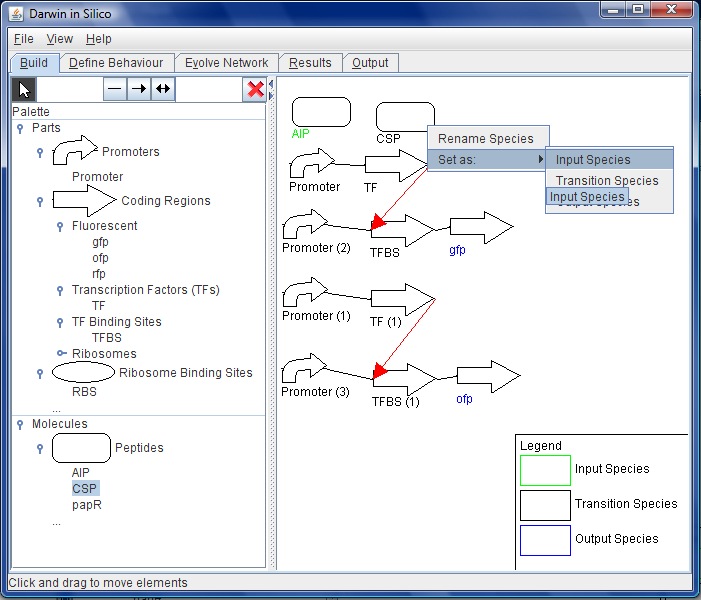
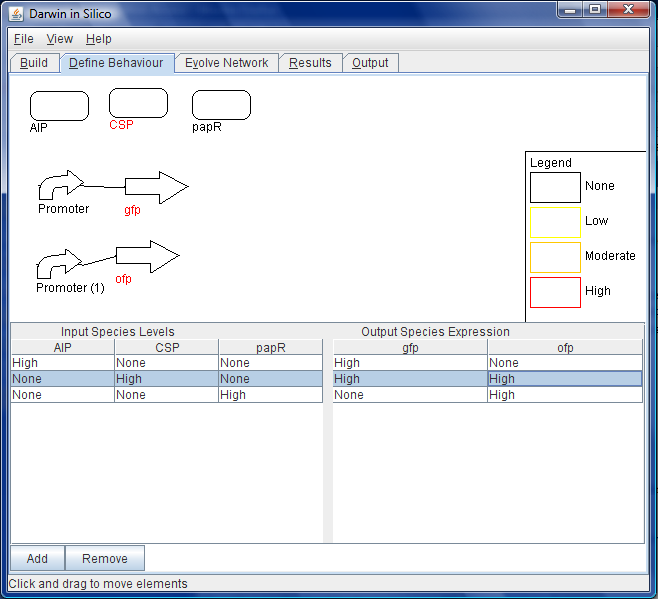
| + | [[Image:Wb-states.png|thumb|200px|right|Figure C: Before moving on to the ''Behaviour'' tab, the inputs to the system and the outputs from the circuit are specified by right clicking on the elements.]] | ||
| + | [[Image:Wb-outputs.png|thumb|200px|right|Figure D: In the ''Behaviour'' tab, the behaviour of the system is specified by a series of tables. In this image, when the input molecule "CSP" is present in high quantities, both GFP and OFP will also be expressed at high levels.]] | ||
| + | |||
| + | Darwin ''in Silico'' (DiS) is the Workbench: the graphical user interface (GUI) for the design of the genetic construct that was [[Team:Newcastle University/Notebook|engineered]] into ''B. subtilis''. The [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] (EA), [[Team:Newcastle University/Parts Repository|parts repository]] (PR) and the [[Team:Newcastle University/Constraints Repository|constraints repository]] (CR) were intended to work behind the scenes of this interface in the design of the construct. | ||
| + | |||
| + | |||
| + | The user can design a starting point for the genetic construct in the centre window of the GUI by dragging and dropping parts into place. All of the two-component genes are available for this process, as are a large number of promoters, ribosome binding sites, transcription factors and sigma factors. These parts are accessed from the parts repository database and built into a preliminary construct. | ||
| + | |||
| + | The constraints repository aids in the building of this construct by not allowing parts that are not compatible to be dropped in place next to each other. Where parts are compatible with each other, constraint models are incorporated into the construct that define the interactions between these. | ||
| + | |||
| + | The user can also map system inputs to system outputs that can be used by the EA. For example, the user could specify that in the presence of a certain set of quorum sensing peptides that a certain fluorescent protein would light up. The user can then run the EA through DiS, leading to the mutation of the hidden layer and the output of the hidden layer model that fulfils the output requirements the user has set. | ||
| + | |||
| + | The combination of these three software components were supposed to communicate through DiS. Each team member was responsible for the construction of their component. Unfortunately, since development of the components proceeded in tandem and the lead developer had to leave the country before the PR, CR and EA were complete, the connections between the components were never incorporated into DiS. The outcome of this was a working build, where the evolutionary algorithm could communicate directly with the parts repository, but not with DiS. Once some more work has been carried out on the various components of the overall project, DiS will be functional. | ||
| + | |||
| + | <html><a name="progress"></html> | ||
| + | ==== DiS: The Ideal and the Reality ==== | ||
| + | |||
| + | It is possible to paint a very rosy-tinted picture of how the user might interact with DiS. '''Bolded items''' have been implemented in the current incarnation of DiS. | ||
| + | |||
| + | # '''The user fires up the workbench by double clicking an icon on the desktop etc.''' | ||
| + | # A nice splash screen appears welcoming them to 'Darwin in Silico: Synthetic Biology Workbench' | ||
| + | # '''The main application window appears.''' If this is the first time the program has run then a wizard may appear asking them to setup configuration details for the rest of the components of the system. If not, then it know where all of these are located by reference to its own internal persistence store of some kind. | ||
| + | # '''The application consists of a working area where a genetic circuit can be specified and a pallete of 'parts' read from the parts repository. These parts have pretty 'BioBrick' type icons''' and can be expanded to give the actual instances of the various part types available in the database. | ||
| + | # The user selects 'New Project' and has to define some initial starting information. They have to enter their name etc and some pre-saved details about them pop up. They modify some information about the ''in silico'' experiment purpose etc. | ||
| + | # '''They are then taken to the blank working area where they can start to define details of the circuit to be evolved. See Figure A.''' | ||
| + | # Somewhere is a menu relating to chassis details. They select ''Bacillus subtilis 168'' from a drop down list or the like. | ||
| + | # Immediately the parts list is constrained by the parts that are available for operation in ''B. subtilis'' 168. | ||
| + | # '''The user begins to add the initial parts to the circuit to be evolved. They know that they want the circuit to essentially respond to the concentration of three (of course may be more or less than this) peptides that are signaling molecules from outside. She select free peptides from the palette at the side and drags them on to the canvas on eat a time where they appear; AIP (a ''Staph aureus'' virulence regulating peptide), CSP (a competence stimulating peptide from ''Streptococcus pneumoniae'') and papR (a virulence controlling peptide from ''Bacillus anthracis'', the causative agent of anthrax). See Figure B.''' | ||
| + | # '''She defines these quantitative levels of these peptides as inputs to the system somehow. ''' | ||
| + | # '''She then selects coding regions from the palette, and navigates to Green Fluorescent protein (gfp) coding region and drags the rectangular icon on to the work area. She repeats this exercise for orange and red fluorescent (ofp and rfp) protein. They automatically appear with a RBS that is optimal for the chosen chassis. She leaves them promoterless for now.''' | ||
| + | # '''She highlights the fluorescent protein coding regions and marks them as system outputs in terms of the absolute levels of the proteins they encode. See Figure C.''' | ||
| + | # '''Next she specifies the functions that specify the behaviour of the outputs for a specified input concentration, again specified by a truth table. In this case she specifies a high initial concentration for AIP and a corresponding high output for gfp and ofp. She assigns zero production to CSP, PapR and rfp.''' | ||
| + | # '''She repeats this exercise for a new set of states. This time she chooses a high initial concentration for PapR and a corresponding high production to rfp and ofp. She assigns a zero production for CSP, AIP and gfp.''' | ||
| + | # '''The process of 13 and 14 is repeated until the desired system behaviour has been fully specified. See Figure D.''' | ||
| + | # '''Next she chooses the chassis location for the fluorescent protein coding regions. She right clicks on them and chooses high copy number plasmid pBD9 (she could have chosen chromosome instead or a range of other vectors)''' | ||
| + | # '''She is almost ready to invoke the evolutionary algorithm. However, she has the option to narrow down the range of parts available to the evolutionary algorithm. In this case she restricts it to all transcription factors and their modifiers and all promoters that operate in the chosen chassis. See Figure E.''' | ||
| + | # She then clicks the 'evolve this' button. Behind the scenes, the model representing this initial set of parts, together with metadata and data about the input/output behaviour is sent to the EA component. | ||
| + | # A dialog box showing the progress of the EA component pops up. | ||
| + | # Once the EA has completed a window pops up showing the ID list of models that meet the fitness function specifications. The user can browse the selection graphically by selection of the relevant model. The graphical selection can be viewed as a conventional pictorial diagram of genetic regulation beloved of biologists or as a graph based view of the model. See Figure F. | ||
| + | # The user is able to view the levels of the different species in the model over time or with varying concentrations of the input signals as line type graphs. | ||
| + | # Ideally, she is able to pick one model tweak it and ask for it to be re-evolved. | ||
| + | # Finally once happy with the model, she clicks on 'model to sequence converter' and the chosen model is sent to the model to sequence converter. A GFF or EMBL formatted record is returned, for each of the genetic elements that require to be synthesised. | ||
| + | <table border="1" width="203"><tr><td valign="top"> | ||
| + | [[Image:Wb-evolve.png|200px]] | ||
| + | <br>Figure E:In the ''Evolve'' tab, the user selects where the circuit should be placed - in a specified vector, or in the chromosome, for example. Then she chooses the parts that should be evolved, and clicks the 'Evolve!' button. | ||
| + | </td><td valign="top"> | ||
| + | [[Image:Wb-results.png|left|200px]] | ||
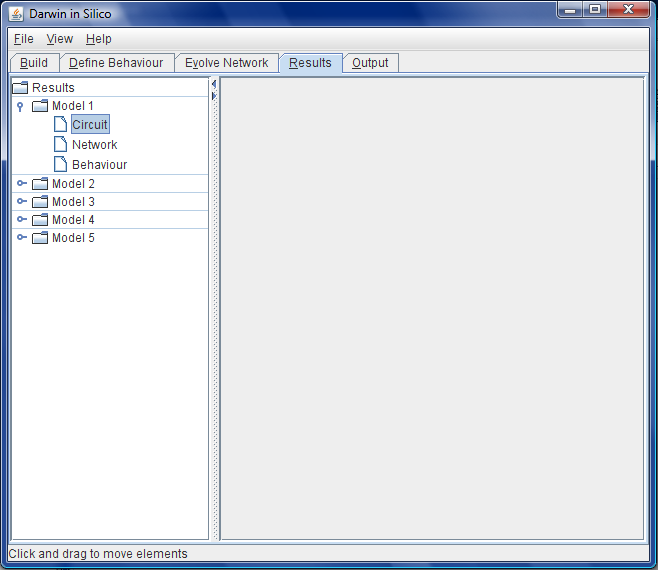
| + | <br>Figure F: Someday, when DiS is hooked up to the EA, the ''Results'' tab will be populated with a series of potential models that the user can save, edit and re-evolve, or export for synthesis and ''in vitro'' biological experiments. | ||
| + | </td></tr></table> | ||
| + | |||
| + | <html><a name="further-reading"></html> | ||
| + | ===Further Reading=== | ||
| + | |||
| + | |||
| + | <html><a name="contributors"></html> | ||
===Contributors=== | ===Contributors=== | ||
Lead: [[Team:Newcastle University/Morgan Taschuk|Morgan Taschuk]] | Lead: [[Team:Newcastle University/Morgan Taschuk|Morgan Taschuk]] | ||
| + | |||
| + | |||
| + | |||
| + | <div id="sidebar"> | ||
| + | {{:Team:Newcastle University/Template:PostItBox | ||
| + | |boxtype=bluebox | ||
| + | |title=Darwin ''in Silico'': Workbench | ||
| + | |detail-text=<ul> | ||
| + | <li>[[Team:Newcastle University/Workbench#aims|Aims and Objectives]] | ||
| + | <li>[[Team:Newcastle University/Workbench#user-stories|User Stories]] | ||
| + | <li>[[Team:Newcastle University/Workbench#outcomes|Outcomes]] | ||
| + | <li>[[Team:Newcastle University/Workbench#progress|DiS: the Ideal and the Reality]] | ||
| + | <li>[[Team:Newcastle University/Workbench#further-reading|Further Reading]] | ||
| + | <li>[[Team:Newcastle University/Workbench#contributors|Contributors]] | ||
| + | </ul> | ||
| + | |link=}} | ||
| + | </div> | ||
Latest revision as of 15:29, 29 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Workbench
Aims and Objectives :
The proposed Synthetic Biology Workbench (cool name needed) will incorporate the functionality of a biological network modeling and simulation program along with spiffy new algorithms that can be used in modern synthetic biology. This will enable users to finetune the expression of models suitable for their purposes, particularly for such fun activities like iGEM competitions.
From a previously created repository of parts and devices, and using the (gentle) guidance of a set of assembly parameters, users will be able to create, edit and piece together biological networks at varying levels of complexity. Users will be able to piece together parts, adding ribosome binding sites, protein coding regions, terminators, etc. until they have exactly the device they desire.
Once the biological circuit is ready in principle, it can be put into practice. The user will be able to request the optimal biological circuit, composed from BioBricks from the repository.
Our objectives are to:
- get a model
- load from SBML and/or CellML
- create using parts and constraints
- show the model
- network view
- basic parts view. Matt suggested being able to view both strands and flip them around.
- take in conditions for synthetic evolution and send models to the evolutionary algorithm
- output the sequence in a standard format
User Stories
Parts Repository
- The program must display a list of all available parts. The program requests a hierarchical list of all part names, part types, and their part ids from the Parts Repository (PR). Upon receiving the list, the program sorts the parts into different categories based on the part type, chooses a display icon for each type, and presents the list of part names with their part type icons to the user.
- The user wishes to change the type of chassis. The program retrieves a list of all chassis names and chassis ids from the PR and presents it to the user.
- The program must display a list of all types of parts. It requests a list of all part types and their part type ids from the PR. Then it displays the list of part names to the user.
- The program must display all of the information about a single part. It gives the part id to the PR and receives a full description of the part in return. It then displays the part information to the user.
- The program must save a CellML model of a part. It gives the part id to the PR and receives a segment of CellML in return. It then saves the CellML to file.
Constraints Repository
- The program must show all possible interactions to a particular part. It passes the part id to the Constraints Repository and receives a list of all possible interaction ids in return. It then retrieves the details of each interaction with the interaction id. It then shows the possible interactions to the user.
Evolutionary Algorithm
- The user finally wants to evolve her model in silico. The program collects the starting model, composed of a series of part ids, and the proposed behaviour in the form of a truth table, and gives it to the Evolutionary Algorithm (EA) for evolution. The program shows a “busy working” screen while the Evolutionary Algorithm computes the best model. When the EA finishes, the program receives the proposed best model(s) and displays them, along with their behaviour, to the user.
Outcome
Darwin in Silico (DiS) is the Workbench: the graphical user interface (GUI) for the design of the genetic construct that was engineered into B. subtilis. The evolutionary algorithm (EA), parts repository (PR) and the constraints repository (CR) were intended to work behind the scenes of this interface in the design of the construct.
The user can design a starting point for the genetic construct in the centre window of the GUI by dragging and dropping parts into place. All of the two-component genes are available for this process, as are a large number of promoters, ribosome binding sites, transcription factors and sigma factors. These parts are accessed from the parts repository database and built into a preliminary construct.
The constraints repository aids in the building of this construct by not allowing parts that are not compatible to be dropped in place next to each other. Where parts are compatible with each other, constraint models are incorporated into the construct that define the interactions between these.
The user can also map system inputs to system outputs that can be used by the EA. For example, the user could specify that in the presence of a certain set of quorum sensing peptides that a certain fluorescent protein would light up. The user can then run the EA through DiS, leading to the mutation of the hidden layer and the output of the hidden layer model that fulfils the output requirements the user has set.
The combination of these three software components were supposed to communicate through DiS. Each team member was responsible for the construction of their component. Unfortunately, since development of the components proceeded in tandem and the lead developer had to leave the country before the PR, CR and EA were complete, the connections between the components were never incorporated into DiS. The outcome of this was a working build, where the evolutionary algorithm could communicate directly with the parts repository, but not with DiS. Once some more work has been carried out on the various components of the overall project, DiS will be functional.
DiS: The Ideal and the Reality
It is possible to paint a very rosy-tinted picture of how the user might interact with DiS. Bolded items have been implemented in the current incarnation of DiS.
- The user fires up the workbench by double clicking an icon on the desktop etc.
- A nice splash screen appears welcoming them to 'Darwin in Silico: Synthetic Biology Workbench'
- The main application window appears. If this is the first time the program has run then a wizard may appear asking them to setup configuration details for the rest of the components of the system. If not, then it know where all of these are located by reference to its own internal persistence store of some kind.
- The application consists of a working area where a genetic circuit can be specified and a pallete of 'parts' read from the parts repository. These parts have pretty 'BioBrick' type icons and can be expanded to give the actual instances of the various part types available in the database.
- The user selects 'New Project' and has to define some initial starting information. They have to enter their name etc and some pre-saved details about them pop up. They modify some information about the in silico experiment purpose etc.
- They are then taken to the blank working area where they can start to define details of the circuit to be evolved. See Figure A.
- Somewhere is a menu relating to chassis details. They select Bacillus subtilis 168 from a drop down list or the like.
- Immediately the parts list is constrained by the parts that are available for operation in B. subtilis 168.
- The user begins to add the initial parts to the circuit to be evolved. They know that they want the circuit to essentially respond to the concentration of three (of course may be more or less than this) peptides that are signaling molecules from outside. She select free peptides from the palette at the side and drags them on to the canvas on eat a time where they appear; AIP (a Staph aureus virulence regulating peptide), CSP (a competence stimulating peptide from Streptococcus pneumoniae) and papR (a virulence controlling peptide from Bacillus anthracis, the causative agent of anthrax). See Figure B.
- She defines these quantitative levels of these peptides as inputs to the system somehow.
- She then selects coding regions from the palette, and navigates to Green Fluorescent protein (gfp) coding region and drags the rectangular icon on to the work area. She repeats this exercise for orange and red fluorescent (ofp and rfp) protein. They automatically appear with a RBS that is optimal for the chosen chassis. She leaves them promoterless for now.
- She highlights the fluorescent protein coding regions and marks them as system outputs in terms of the absolute levels of the proteins they encode. See Figure C.
- Next she specifies the functions that specify the behaviour of the outputs for a specified input concentration, again specified by a truth table. In this case she specifies a high initial concentration for AIP and a corresponding high output for gfp and ofp. She assigns zero production to CSP, PapR and rfp.
- She repeats this exercise for a new set of states. This time she chooses a high initial concentration for PapR and a corresponding high production to rfp and ofp. She assigns a zero production for CSP, AIP and gfp.
- The process of 13 and 14 is repeated until the desired system behaviour has been fully specified. See Figure D.
- Next she chooses the chassis location for the fluorescent protein coding regions. She right clicks on them and chooses high copy number plasmid pBD9 (she could have chosen chromosome instead or a range of other vectors)
- She is almost ready to invoke the evolutionary algorithm. However, she has the option to narrow down the range of parts available to the evolutionary algorithm. In this case she restricts it to all transcription factors and their modifiers and all promoters that operate in the chosen chassis. See Figure E.
- She then clicks the 'evolve this' button. Behind the scenes, the model representing this initial set of parts, together with metadata and data about the input/output behaviour is sent to the EA component.
- A dialog box showing the progress of the EA component pops up.
- Once the EA has completed a window pops up showing the ID list of models that meet the fitness function specifications. The user can browse the selection graphically by selection of the relevant model. The graphical selection can be viewed as a conventional pictorial diagram of genetic regulation beloved of biologists or as a graph based view of the model. See Figure F.
- The user is able to view the levels of the different species in the model over time or with varying concentrations of the input signals as line type graphs.
- Ideally, she is able to pick one model tweak it and ask for it to be re-evolved.
- Finally once happy with the model, she clicks on 'model to sequence converter' and the chosen model is sent to the model to sequence converter. A GFF or EMBL formatted record is returned, for each of the genetic elements that require to be synthesised.
Further Reading
Contributors
Lead: Morgan Taschuk
 "
"