|
|
| (23 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{:Team:Newcastle University/Header}} | | {{:Team:Newcastle University/Header}} |
| - | {{:Team:Newcastle University/Template:UnderTheHome|page-title=[[Team:Newcastle University/Parallel Evolution|Approach]]}} | + | {{:Team:Newcastle University/Template:UnderTheHome|page-title=[[Team:Newcastle University/Original Aims|Original Aims]]}} |
| | + | == Original Aims == |
| | | | |
| - | == Parallel Evolution ==
| + | Bioinformatics is essential to modern biology, but is often considered as merely a tool, like a word processor or a spreadsheet, rather than an important approach to understanding and solving biological problems. |
| | | | |
| - | There standard practice in biology today, synthetic or not, is to treat bioinformatics as any other tool to achieve an end. A problem is determined, and the biologist works out a possible avenue of exploration. Sometimes, this involves the use of bioinformatics tools, such as BLAST searches or phenotypic trees. Once the method of exploration is established, the biologist rarely goes back to bioinformatics approaches to analyze her results.
| + | We consider bioinformatics to be not only a tool in biology, but as a viable method for exploring a problem. Wet-lab biology is expensive in terms of time, money, and manpower. A single bioinformatician can investigate a given problem with no equipment other than a computer, and run many iterations of the same experiment within seconds, rather than the weeks that the labs may take. |
| | | | |
| - | The concept of parallel evolution treats bioinformatics not only as a tool in biology, but as a viable but limited method for exploring a problem. Wet-lab biology is expensive in terms of time, money, and manpower. A single bioinformatician can test the same situation with no equipment other than a computer, and run many iterations of the same experiment within seconds, rather than the weeks that the labs may take.
| + | However, the bioinformatics is only as good as its data and simulation power. The field expands daily with new information, all of which must be incorporated into a simulation in order for it to give useful results. Much of bioinformatics is backed by hard data from wet labs. Running an experiment 1,000 times is only useful if you know what all the variables are, and what they should be. |
| | | | |
| - | However, the bioinformatics is only as good as its simulation. The field expands daily with new information, all of which must be incorporated into the simulation in order for it to give useful results. Much of bioinformatics is backed by hard data from wetlabs. Running an experiment 1000 times is only useful if you know what all the variable are, and what they should be.
| + | The most sensible approach to the biological investigations is to take advantahe both of of these different yet complementary methods - bioinformatics and wet lab experimentation - taking advantage of the strengths of each, while hopefully minimizing the limitations. |
| | + | ==BugBuster== |
| | + | We aimed to develop a diagnostic biosensor for detecting pathogens. We wanted this to be cheaply and readily available for deployment in areas with limited medical resources such as refrigeration and sophisticated laboratories. We chose to use ''Bacillus subtilis'' as a method of delivery due to its ability to sporulate. The sensor bacteria could be dried down as spores, which are extremely resilient to environmental conditions, and could be rehydrated as required. |
| | | | |
| - | The most sensible approach to the problem of these different yet complementary methods is to play one off of the other, taking advantage of the strengths while minimizing the limitations.
| + | Our aim was to develop a strain of [[Team:Newcastle University/Bacillus subtilis|''Bacillis subtilis 168'']] with the ability to detect four Gram-positive pathogens through their extracellularly-secreted quorum-sensing peptides, and to indicate through reporter genes linked to the receptor-ligand cascade which of these peptides are in its growth medium. |
| | | | |
| | + | Gram-positive bacteria communicate using quorum communication peptides. Research has shown that they are extremely strain-specific. ''B. subtilis'' has a range of genes that enable detection of these peptides. The issue that we had to overcome is the fact that we have more quorum-sensing peptides to detect than fluorescent proteins to show their presence. The output of the system is followed by analysing expression of fluorescent proteins such as mCherry, GFP, CFP and YFP. |
| | | | |
| - | == Drylab Approach ==
| + | Mapping multiple inputs to three output states is a multiplexing problem. The design of the genetic circuitry to do this is non-trivial and is not feasible manually. We therefore chose to use a biological implementation of an artificial neural network (ANN). Our team members wrote, designed and implemented a complete suite of tools that allowed the design and simulation of regulatory networks. An essential part of the approach was the use of computational evolution to design circuits with predictable behaviour even when the details of the required topology are unknown in advance. We used the modelling language CellML[http://www.cellml.org/] because of its ability to easily model virtual parts that can be easily assembled into a circuit which can be simulated. We aimed to translate this model into a sequence that could be implemented as one or more BioBricks. |
| | | | |
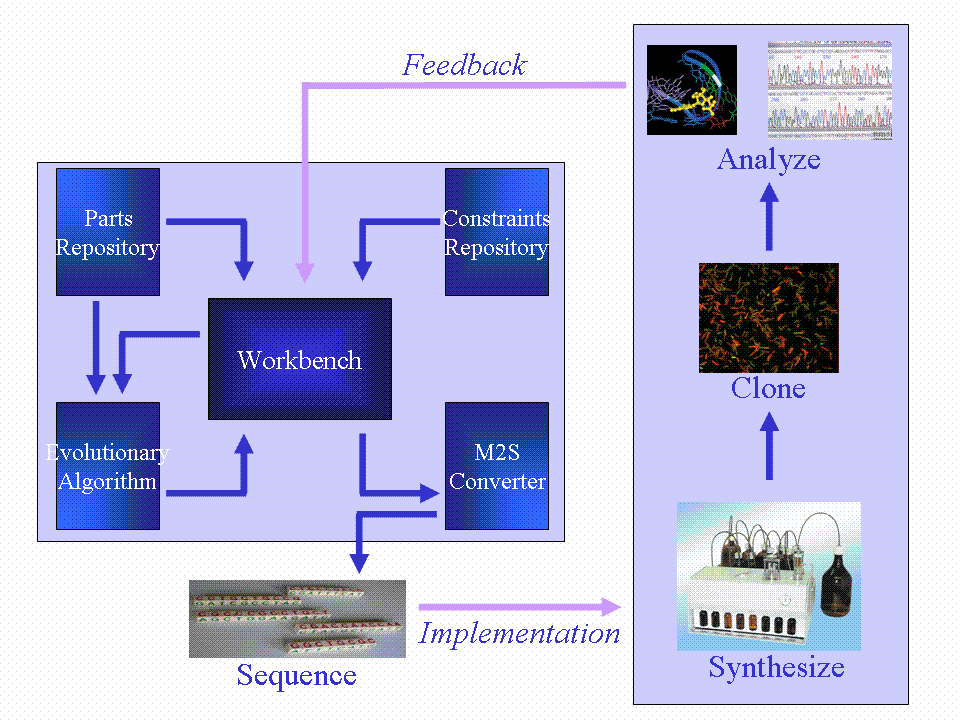
| - | The two techniques we decided to use in our bioinformatics approach were neural networks and genetic algorithms. Computer science has consistently looked to Mother Nature for inspiration, and the results are best exemplified with these two techniques.
| + | [[Image:Overview.GIF|500px|center]] |
| | | | |
| - | [[Image:Neuron.png|right|thumbnail|A quick sketch of the anatomy and connection of neurons]]
| + | <center>An overview of our complete system.</center> |
| - | Neural networks are a method of machine learning based on neurons in the brain. In a neuron, signals are received at the dendrites. If the combined triggering impulse is higher than a certain threshold, the neuron fires. The impulse travels down the cell membrane of the axon and is transmitted via the axon terminators to the dendrites of the next neuron. When the neuronic pathway is traveled often, the connections are reinforced. The axon of the preceeding neuron and the dendrites of the subsequent neuron grow to be more highly connected. It is in this way that we learn. and why we can change behaviour patterns once they are established. Forcing the impulse to take another route results in an altered behaviour or thought.
| + | |
| | | | |
| - | This same approach is used to teach a computer how to 'think' about a problem. The neurons in the system become nodes. The nodes are arranged in several layers. "Impulses" begin at the input layer, travel through any number of hidden layers, and terminate at the output layer. The output layer gives the computer its learned behaviour.
| + | A visual output was chosen for detection of pathogens, as this allows for rapid detection without a requirement for specialist equipment. The aim was to have different fluorescent protein outputs turned on by the presence of different pathogenic bacteria, and different combinations of these bacteria. In line with the neural network concept we wanted to map numerous inputs to limited outputs. Initially, we aimed to detect four pathogens, and we wanted different outputs for each of these and each combination of these. |
| | | | |
| - | Nodes can be highly interconnected. The connections themselves have a weight, which corresponds to the number of axon-dendrite connections in neurons. Nodes that are connected with a heigher weight are more likely to fire.
| + | We aimed to produce a [[Team:Newcastle University/Workbench|workbench]] that incorporates a [[Team:Newcastle University/Parts Repository|parts repository]], [[Team:Newcastle University/Constraints Repository|constraints repository]] and an [[Team:Newcastle University/Evolutionary Algorithm|evolutionary algorithm]] (EA). The EA takes input from the parts repository and constraints repository and evolves a neural network, using the CellML modelling language to carry out simulations of the network behaviour, which can be assessed for fitness (how closely the desired behaviour is reproduced). The fittest model can then be used to generate a DNA sequence implementing the neural network ''in vivo''. This DNA sequence can then be synthesized and cloned into the ''B. subtilis'' chassis. One of our outcomes should be a range of neural network node BioBrick devices which can be combined to form the ''in vivo'' neural network. |
| | | | |
| - | Neural networks must be trained in order to function properly. Untrained neural networks are like newborn babies: cute, but unable to perform any higher functions. They must be taught.
| + | In keeping with the neural network model, our bacterial circuit would have three layers of complexity. The plan can be summed up as follows: |
| | | | |
| - | ==Wetlab Approach==
| + | * The input layer to the bacterial neural network is represented by the two-component genes, activated by the peptides from the four gram-positive pathogens. Each peptide represents a node. |
| | + | * The hidden layer is represented by an assortment of different transcription factors. |
| | + | * The output layer is represented by three fluorescent proteins; GFP, YFP and mCherry. |
| | | | |
| - | Newcastle’s 2008 iGEM team is aiming to transform plasmids grown in ''Escherichia coli'' into ''Bacillis subtilis'' (a gram-positive bacterium), with the aim of integrating this stably into the ''B. subtilis'' chromosomal DNA. By doing this, we hope to create an organism with the ability to detect a range of extracellularly secreted quorum-sensing peptides, and indicate through reporter genes linked to the receptor-ligand cascade which of these are in its growth medium.
| |
| | | | |
| - | ===Advantages of using B. Subtilis===
| + | The biologist specifies the inputs and the outputs. In the presence of a particular profile of peptides, a specific fluorescent protein, or combination of proteins, will be expressed. |
| - | There are several reasons why we chose to use ''B.subtilis'', as supposed to E.coli as our chassis.
| + | |
| | | | |
| - | ====Pathogenesis====
| + | This was the starting point from which the construct was designed. |
| - | ''B.subtilis'' is categorised under Biohazard Level 1- i.e it is a minimum risk bacteria. As a result of not being considered a human pathogen, the only necessary precautions to be implicated include using gloves, hand-washing, and autoclaving equipment. This saves time and money.
| + | |
| - | | + | |
| - | ====Gram-Positive====
| + | |
| - | The Gram stain technique is based on the ability of a bacterial cell wall to retain a crystal violet dye- allowing us to differentiate between two major cell wall types:
| + | |
| - | | + | |
| - | * '''Gram negative''' cell walls have a lower percentage of peptidoglycan. Gram-negative bacteria also have an additional outer membrane containing lipids, separated from the cell wall by the periplasmic space.
| + | |
| - | * '''Gram positive''' cell walls contain a relatively large amount of peptidoglycan (up to 90%) but lower lipid content. This stains purple upon addition of crystal violet dye, whereas the wall of a gram negative bacteria will only stain pink.
| + | |
| - | | + | |
| - | After decolorisation, Gram-positive cells remain purple and Gram-negative cells lose their purple colour. When counterstain is applied, Gram-negative bacteria turn red.
| + | |
| - | | + | |
| - | For a tutorial on Gram staining see:
| + | |
| - | http://www.ncl.ac.-uk/dental/oralbiol/oralenv/tutorials/gramstain.htm
| + | |
| - | | + | |
| - | ''B. subtilis'' is a gram-positive bacterium and therefore contains only a single membrane. Consequently, they absorb substances better than ''E.coli'', and this is ideal for our project.
| + | |
| - | | + | |
| - | ====Transformation====
| + | |
| - | ''B. subtilis'' has the ability to take up exogenous DNA from the surrounding medium and integrate it in their genome. This natural competency can be tightly regulated and induced in the lab by nutritional shortages using starvation medium.
| + | |
| - | | + | |
| - | This makes transformation easier, as competence comes naturally to B.subtilis.
| + | |
| - | | + | |
| - | ====Sporulation====
| + | |
| - | During times of nutritional stress, ''B.subtilis'' cells produce an endospore which enables it to withstand extreme conditions such as high temperatures and salt concentrations. The purpose of this is to allow the bacterium to survive in the unfavourable environment until conditions return back to normal. Prior to the decision to produce the spore the bacterium may take up DNA from the surrounding medium.
| + | |
| - | | + | |
| - | This means that ''B.subtilis'' can be easily stored and transported without damaging the cells and their ability to differentiate.
| + | |
| - | | + | |
| - | ===The Centre for Bacterial Cell Biology===
| + | |
| - | In addition, Newcastle is lucky enough to have The Centre for Bacterial Cell Biology (CBCB), part of the Institute for Cell and Molecular Biosciences (ICaMB) at Newcastle University, on its doorstep. As a result, we can make use of the expertise of the researchers based at the centre- which includes some of our advisors
| + | |
| - | | + | |
| - | Although ''B. subtilis'' has many clear benefits in its usage, gram-positive bacteria are generally relatively poorly characterised and consequently can be unpredictable to work with. Nevertheless, ''B. subtilis'' is one of the best known gram-positives. This page intends to cover some issues about working with this bacterium.
| + | |
| - | | + | |
| - | In this project, we used ''B. subtilis'' 168 (Bs168). This is a strain which lacks the trp operon and subsequently must have tryptophan present in its growth medium as it cannot synthesise this essential amino acid. The inability to synthesise tryptophan means that Bs168 is nutrient-inducible (a factor that can be exploited when introducing exogenous DNA).
| + | |
| - | | + | |
| - | How using a B. subtilis vector is different to using an E. coli vector
| + | |
| - | | + | |
| - | ===Genetic Transformation===
| + | |
| - | | + | |
| - | The transformation procedures in ''E. coli'' and ''B. subtilis'' are very different. The ''E. coli'' strains we were working with (TOP10 and DH5α) were already competent to take up exogenous DNA, and therefore require only basic preparation before transformation. This briefly consists of adding the DNA to the cells and incubating them on ice, then heat shocking and adding LB before incubating again at 37°C.
| + | |
| - | | + | |
| - | In contrast, ''B. subtilis'' cells must be induced to take up foreign DNA. Research has shown that when ''B. subtilis'' “encounters nutrient limitations and enters the stationary growth phase, it can engage in multiple adaptation strategies such as…uptake of exogenous DNA (competence)” (1). It is therefore important to carefully control the growth phase and nutrient availability in the ''B. subtilis'' culture prior to transformation. We used the protocol below to achieve this.
| + | |
| - | | + | |
| - | One of the main advantages of using ''B. subtilis'' is that it is naturally competent for transformation. When bacteria are in the stationary growth phase and are nutrient-starved, the cells begin to transcribe high levels of a tetramic protein called ComK, a transcription factor that activates over 100 genes (“including those essential for DNA binding and uptake” (2). The competency of the cells to take up exogenous DNA is transient, and therefore observing correct timings in the transformation protocol is very important in increasing the population of competent cells.
| + | |
| - | | + | |
| - | ====Protocol for Genetic Transformation into B. subtilis====
| + | |
| - | | + | |
| - | Prepare two conical shake flasks of the following medium:
| + | |
| - | * 10mL SMM
| + | |
| - | ** 125 µl sol E (40% glucose)
| + | |
| - | ** 100 µl tryptophan
| + | |
| - | ** 60 µl sol F
| + | |
| - | ** 10 µl casamino acid (CAA)
| + | |
| - | ** 5µl ammonium iron
| + | |
| - | * To one flask add 2mL liquid culture cells.
| + | |
| - | * Incubate both flasks at 37°C overnight. (''It is important that the flask without cells in is kept at 37°C also as this is the dilution medium and must be pre-warmed at the same temperature as the culture to avoid temperature stress of the cells.'')
| + | |
| - | * Add 500 µl overnight culture to the pre-warmed dilution medium and leave to incubate at 37 °C for 3 hours. (''This allows the cells to enter the stationary growth phase.'')
| + | |
| - | * Add 10mL SMM, 125 µl sol E and 60 µl sol F to the dilute culture and leave to incubate at 37°C for a further 2 hours. (''This starves the cells of amino acids such as tryptophan and CAA.'')
| + | |
| - | Transfer 10µl plasmid to be transformed into a 2mL tube. Add 400µl nutrient-starved cell culture and mix by inverting the tube five times.
| + | |
| - | * Incubate at 37°C whilst shaking for 1 hour. (''The tubes must be laid on their side to properly aerate the medium. To do this, tape the tubes to the rack and lay it on its side to prevent the tubes falling out whilst shaking.'')
| + | |
| - | * Centrifuge the tube at 13,000g for 2 minutes to pellet the cells.
| + | |
| - | * Use a pipette to remove 300µl supernatant.
| + | |
| - | * Pipette the remaining liquid and cells up and down to resuspend and plate immediately onto agar. (''The pellet self-resuspends very quickly. If it has already resuspended before the supernatant is removed, the tube must be spun again to avoid loss of cells.'')
| + | |
| - | | + | |
| - | ===Temperature Sensitivity===
| + | |
| - | | + | |
| - | Unlike E. coli, B. subtilis is very temperature sensitive and sudden changes in temperature can be detrimental to it. When culturing, the cells should be kept at their 37°C optimum. We cultured all our colonies in a 37°C culture room. This way we could perform all pipetting and diluting of cells without changing their temperature.
| + | |
| - | | + | |
| - | ===Oxygen Requirements===
| + | |
| - | | + | |
| - | B. subtilis is a strict aerobe and also requires much more oxygen than E. coli. If they become anoxic they will quickly die. Therefore it is important for culture media to always be kept shaking whilst the cells are growing to constantly provide them with oxygen. Allowing the broth to rest even for short periods of time can result in a lower transformation efficiency. All pipetting should be performed as quickly as possible to reduce the amount of time when the culture media are still.
| + | |
| - | | + | |
| - | When making overnight cultures, conical shake flasks should preferably be used. The foam plug in the neck of the flasks allows exchange of air between the interior and exterior of the flask. Capped glass tubes can also be used as the caps are loose fitting and also allow air passage. Screw-cap plastic tubes should never be used as these are airtight. This is important for all overnight bacterial cultures but particularly for B. subtilis as it cannot survive anoxic conditions.
| + | |
| - | | + | |
| - | ===Problems Encountered when Working with B. subtilis===
| + | |
| - | When we began work on B.subtilis we knew very little about the organism and therefore made some basic errors. For one transformation we used screw-cap plastic tubes, which significantly reduced both the number and size of the colonies on agar. Leaving them to grow for a further 6 hours allowed them to grow large enough for colony stabs to be taken.
| + | |
| - | | + | |
| - | ===OTHER STUFF..===
| + | |
| - | * which vectors work best with it
| + | |
| - | * plasmid diagrams/ restriction maps
| + | |
| - | * ??
| + | |
| - | | + | |
| - | ===References===
| + | |
| - | - Veening, J.-W., Igoshin, O. A., Eijlander, R. T., Nijland, R., Hamoen, L. W., Kuipers, O. P. (Apr 2008) “Transient Heterogeneity in Extracellular Protease Production by Bacillus subtilis” Molecular Systems Biology 4: 184
| + | |
| - | - Smits, W. K., Kuipers, O. P., Veening, J.-W. (2006) “Phenotypic Variation in Bacteria: the Role of Feedback Regulation” Nature 4: 259-271
| + | |
| - | | + | |
| - | ===Ribosome Binding Sites (RBS) for B. subtilis===
| + | |
| - | | + | |
| - | Perfect Bacillus RBS' + atg:
| + | |
| - | | + | |
| - | TAAGGAGGAACTACTATG
| + | |
| - | | + | |
| - | See Vellanoweth and Rabinowitz (Mol Micro., 1992, 6 1105-1114) and Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat. Genet 2002 May;31(1):69-73
| + | |
| - | | + | |
| - | ===Promoters===
| + | |
| - | | + | |
| - | All promoters below contain an RBS and the minimal promoter sequence is not known for all of these.
| + | |
| - | | + | |
| - | Inducible promoters in B.subtilis: Phyper_spank:
| + | |
| - | | + | |
| - | Hy_spank (IPTG inducible if LacI is present, otherwise very strong constitutive; range between 0 and 1 mM IPTG):
| + | |
| - | <pre>
| + | |
| - | GACTCTCTAGCTTGAGGCATCAAATAAAACGAAAGGCTCAGTCGAAAGACTGGGCCTTTC
| + | |
| - | GTTTTATCTGTTGTTTGTCGGTGAACGCTCTCCTGAGTAGGACAAATCCGCCGCTCTAGC
| + | |
| - | TAAGCAGAAGGCCATCCTGACGGATGGCCTTTTTGCGTTTCTACAAACTCTTGTTAACTC
| + | |
| - | TAGAGCTGCCTGCCGCGTTTCGGTGATGAAGATCTTCCCGATGATTAATTAATTCAGAAC
| + | |
| - | GCTCGGTTGCCGCCGGGCGTTTTTTATGCAGCAATGGCAAGAACGTTGCTCGAGGGTAAA
| + | |
| - | TGTGAGCACTCACAATTCATTTTGCAAAAGTTGTTGACTTTATCTACAAGGTGTGGCATA
| + | |
| - | ATGTGTGTAATTGTGAGCGGATAACAATTTAAGGAGGAACTACTATG
| + | |
| - | </pre>
| + | |
| - | | + | |
| - | See e.g.: Quisel, J., Burkholder, W.F., and Grossman, A.D. J Bacteriol. 2001 183 6573-8
| + | |
| - | | + | |
| - | | + | |
| - | (image from David Rudner, unpublished).
| + | |
| - | | + | |
| - | Pxyl (xylose inducible, range between 0 and 2 % xylose, depends on relief from XylR repressor):
| + | |
| - | <pre>
| + | |
| - | TCGGATCTTCATGAAAAACTAAAAAAAATATTGAAAATACTGATGAGGTTATTTAAGATT
| + | |
| - | AAAATAAGTTAGTTTGTTTGGGCAACAAACTAATGTGCAACTTACTTACAATATGACATA
| + | |
| - | AAATGCATCTAGAAAGGAGATTCCTAGGATGGGTACTAAGGAGGAACTACTATG
| + | |
| - | </pre>
| + | |
| - | | + | |
| - | See e.g. Hastrup, S. (1988) Analysis of the Bacillus subtilis xylose region. In Genetics and Biotechnology of Bacilli. Ganesan, A.T., and Hoch, J.A. (eds). New York: Academic Press, pp. 79–83.
| + | |
| - |
| + | |
| - | PspaS (subtilin inducible, range between 0 and 10%, supernatant of a subtilin producing strain; requires the SpaRK 2-component system)
| + | |
| - | <pre>
| + | |
| - | GATCTTAAAAAAAGGAAAAAATTGATAAAATCTTGATATTTGTCTGTTACTATTTAGGTA
| + | |
| - | TTGAAAGGAGGTGACCACCATG
| + | |
| - | </pre>
| + | |
| - | | + | |
| - | Bongers et al, 2005 Applied and Environmental Microbiology
| + | |
| - | | + | |
| - | ===Constitutive promoters in B. subtilis:===
| + | |
| - | | + | |
| - | Low level: PzapA:
| + | |
| - | <pre>
| + | |
| - | CAGCAGGCTGTACGGAATGGTTTTAATCAGGTTTCGGGCAATCTCGATGATTTGCGGATC
| + | |
| - | TTTTAAATCTTTAGAATCCTTAACACCGAGTTCCTTCATTAAAGGAAGCATGGTTTTGTC
| + | |
| - | GACGTACGCGCATACAACCGTCATTGGGCCAAAGTAATCTCCGGTTCCGACTTCGTCAGA
| + | |
| - | ACCGATAACAGACATTCCGGCGATATCTGCCGGAGGGGCATAGCGGGGATCAGCCGGCTT
| + | |
| - | TTTGGCCGTTTTCTTCTTTTCCTGAGGCTCTGCTGTTCCCCAGCGCGCGGATTCTGCCGC
| + | |
| - | AGCGTTTTTTCCTTGAAACAAGACCTTACCTGATTGATATGCTGTAATGGTACAGCCGGG
| + | |
| - | CGGTTTTGCCTGAAAAACGGCTCCCTGCGGAACAGAGGCTGTAAGTGAACCGCTGTACGT
| + | |
| - | CATTTTCATTTGGTCAATAGCAGACAACGATACTTTTATCACTGAATGGGACACGTAATA
| + | |
| - | ATCTCCTTTTTTTACACTTTTCGCTGTATATACCAGTGTATCATAACAGCGGGAGGCTCG
| + | |
| - | TCTTTCCATTCATTTAATAAACGTGTTATGATAAGAACTAGGATTCTCGCGGTAAGGAGG
| + | |
| - | AACTACTATG
| + | |
| - | </pre>
| + | |
| - | Medium level: PftsW:
| + | |
| - | <pre>
| + | |
| - | CTGCGAAAAGCGGCGTATCCGGCGGGATAATGGGCATTTCGCCTTACGATTTCGGGATCG
| + | |
| - | GCATATTCGGCCCCGCATTAGACGAAAAAGGGAACAGCATTGCAGGTGTAAAGCTTTTGG
| + | |
| - | AAATAATGTCTGAGATGTACAGGCTGAGTATTTTTTAATTTATGTCATATGCTTAAATCC
| + | |
| - | TCTTGCATTTTCTGTTGATACCCTTTATGATAAATAGAAGAATTAGGTACTCGCCTGGGG
| + | |
| - | AACGGAGGGATACTTTTGGCTTCAGAGATGATAGTTGACCATCGGCAAAAAGCTTTTGAA
| + | |
| - | CTCTTAAAAGTGGACGCTGAGAAGATTTTGAAGCTGATCCGAGTACAAATGGATAACTTA
| + | |
| - | ACGATGCCTCAATGTCCTCTATATGAAGAGGTTTTAGATACTCAAATGTTCGGGCTTTCG
| + | |
| - | AGGGAAATCGATTTTGCTGTCCGCCTGGGATTGGTTGATGAAAAAGACGGTAAAGACCTT
| + | |
| - | TTATACACATTGGAGCGCGAATTGTCTGCTTTGCATGACGCGTTTACAGCTAAATAAATG
| + | |
| - | ATAAAACTCAAACTTATTAACAGTTTGGGTTTTTTTATAACCGCTATTTTTCTCTCATCT
| + | |
| - | CATAAAAGACGGTCTTTTTTTACACATTCCTTCCGAATCGTATAGAGATTCTTCGTCTCG
| + | |
| - | TTTGATAAATTGTAGTAAAATAAAAAAGATAAATACATAAAAACCATAGATAATGGAAGT
| + | |
| - | TAGAAGCTAAGGAGGAACTACTATG
| + | |
| - | </pre>
| + | |
| - | High level: Pveg
| + | |
| - | <pre>
| + | |
| - | GCGTACAGACATTCTAAGCACTTTGTTGAAACAGTTCAAACAGCCCGAAAAAAGATCCCT
| + | |
| - | CACTTAGATCAGCTTGTTATTTTTGCGGGGGCCTGCCAATCCCATTTTGAATCACTCATC
| + | |
| - | AGAGCGGGTGCGAATTTTGCAAGTTCACCGTCAAGAGTCAATATTCATGCGCTTGATCCG
| + | |
| - | GTATATATCGTCGCGAAGATCAGCTTTACGCCGTTTATGGAACGGATTAATGTATGGGAA
| + | |
| - | GTGCTCCGTAATACGCTGACAAGAGAGAAAGGGCTTGGAGGTATTGAAACAAGAGGAGTT
| + | |
| - | CTGAGAATTGGTATGCCTTATAAGTCCAATTAACAGTTGAAAACCTGCATAGGAGAGCTA
| + | |
| - | TGCGGGTTTTTTATTTTACATAATGATACATAATTTACCGAAACTTGCGGAACATAATTG
| + | |
| - | AGGAATCATAGAATTTTGTCAAAATAATTTTATTGACAACGTCTTATTAACGTTGATATA
| + | |
| - | ATTTAAATTTTATTTGACAAAAATGGGCTCGTGTTGTACAATAAATGTTAAGAGGAGGAA
| + | |
| - | CTACTATG
| + | |
| - | </pre>
| + | |
| - | Bacillus Terminator sequences:
| + | |
| - | | + | |
| - | Bacillus terminators:
| + | |
| - | | + | |
| - | rrnO:
| + | |
| - | <pre>
| + | |
| - | TAGGACGCCGCCAAGCCAGCTTAAACCCAGCTCAATGAGCTGGGTTTTTTGTTTGTTAAA
| + | |
| - | AATGAAGAAGAAACTGTGAAGCGTATTTA
| + | |
| - | </pre>
| + | |
| - | rrnA:
| + | |
| - | <pre>
| + | |
| - | TAAAATCTAAGACATATCATGATTTATAAAAACAAAAAAACCTTGCAGGATATGCAAGGC
| + | |
| - | TTCTGGATGACCCGTACGGGATTCGAAC
| + | |
| - | </pre>
| + | |
| - | | + | |
| - | ==Links==
| + | |
| - | | + | |
| - | [http://www.ncl.ac.uk/camb/cbcb/ The Centre for Bacterial Cell Biology]
| + | |
| - | | + | |
| - | The Institute for Cell and Molecular Biosciences at Newcastle University: http://www.ncl.ac.uk/camb/
| + | |
| - | | + | |
| - | http://www.ncl.ac.uk/camb/staff/profile/colin.harwood
| + | |
| - | | + | |
| - | http://www.ncl.ac.uk/camb/staff/profile/l.hamoen
| + | |
| - | | + | |
| - | http://www.ncl.ac.uk/camb/staff/profile/j.w.veening
| + | |
| - | | + | |
| - | http://www.ncl.ac.uk/camb/staff/profile/jeff.errington
| + | |
| - | http://www.staff.ncl.ac.uk/j.s.hallinan/
| + | |
| - | http://www.ncl.ac.uk/camb/staff/profile/anil.wipat
| + | |
| - | http://www.cs.ncl.ac.uk/people/matthew.pocock
| + | |
| - | School of computing science: http://www.cs.ncl.ac.uk/
| + | |
Bioinformatics is essential to modern biology, but is often considered as merely a tool, like a word processor or a spreadsheet, rather than an important approach to understanding and solving biological problems.
We consider bioinformatics to be not only a tool in biology, but as a viable method for exploring a problem. Wet-lab biology is expensive in terms of time, money, and manpower. A single bioinformatician can investigate a given problem with no equipment other than a computer, and run many iterations of the same experiment within seconds, rather than the weeks that the labs may take.
However, the bioinformatics is only as good as its data and simulation power. The field expands daily with new information, all of which must be incorporated into a simulation in order for it to give useful results. Much of bioinformatics is backed by hard data from wet labs. Running an experiment 1,000 times is only useful if you know what all the variables are, and what they should be.
The most sensible approach to the biological investigations is to take advantahe both of of these different yet complementary methods - bioinformatics and wet lab experimentation - taking advantage of the strengths of each, while hopefully minimizing the limitations.
We aimed to develop a diagnostic biosensor for detecting pathogens. We wanted this to be cheaply and readily available for deployment in areas with limited medical resources such as refrigeration and sophisticated laboratories. We chose to use Bacillus subtilis as a method of delivery due to its ability to sporulate. The sensor bacteria could be dried down as spores, which are extremely resilient to environmental conditions, and could be rehydrated as required.
Gram-positive bacteria communicate using quorum communication peptides. Research has shown that they are extremely strain-specific. B. subtilis has a range of genes that enable detection of these peptides. The issue that we had to overcome is the fact that we have more quorum-sensing peptides to detect than fluorescent proteins to show their presence. The output of the system is followed by analysing expression of fluorescent proteins such as mCherry, GFP, CFP and YFP.
Mapping multiple inputs to three output states is a multiplexing problem. The design of the genetic circuitry to do this is non-trivial and is not feasible manually. We therefore chose to use a biological implementation of an artificial neural network (ANN). Our team members wrote, designed and implemented a complete suite of tools that allowed the design and simulation of regulatory networks. An essential part of the approach was the use of computational evolution to design circuits with predictable behaviour even when the details of the required topology are unknown in advance. We used the modelling language CellML[http://www.cellml.org/] because of its ability to easily model virtual parts that can be easily assembled into a circuit which can be simulated. We aimed to translate this model into a sequence that could be implemented as one or more BioBricks.
A visual output was chosen for detection of pathogens, as this allows for rapid detection without a requirement for specialist equipment. The aim was to have different fluorescent protein outputs turned on by the presence of different pathogenic bacteria, and different combinations of these bacteria. In line with the neural network concept we wanted to map numerous inputs to limited outputs. Initially, we aimed to detect four pathogens, and we wanted different outputs for each of these and each combination of these.
In keeping with the neural network model, our bacterial circuit would have three layers of complexity. The plan can be summed up as follows:
This was the starting point from which the construct was designed.
 "
"