Team:Imperial College/B.subtilis
From 2008.igem.org
| Line 3: | Line 3: | ||
{{Imperial/Box2|Working With ''B. subtilis''|Throughout this summer we worked with ''B. subtilis'' as a chassis. Although this chassis has great potential, it is not commonly used and very few ''B. subtilis'' BioBricks are available. We aimed to increase the accessibility of ''B. subtilis'' as a chassis. The major results of our work are summarised on this page. | {{Imperial/Box2|Working With ''B. subtilis''|Throughout this summer we worked with ''B. subtilis'' as a chassis. Although this chassis has great potential, it is not commonly used and very few ''B. subtilis'' BioBricks are available. We aimed to increase the accessibility of ''B. subtilis'' as a chassis. The major results of our work are summarised on this page. | ||
}} | }} | ||
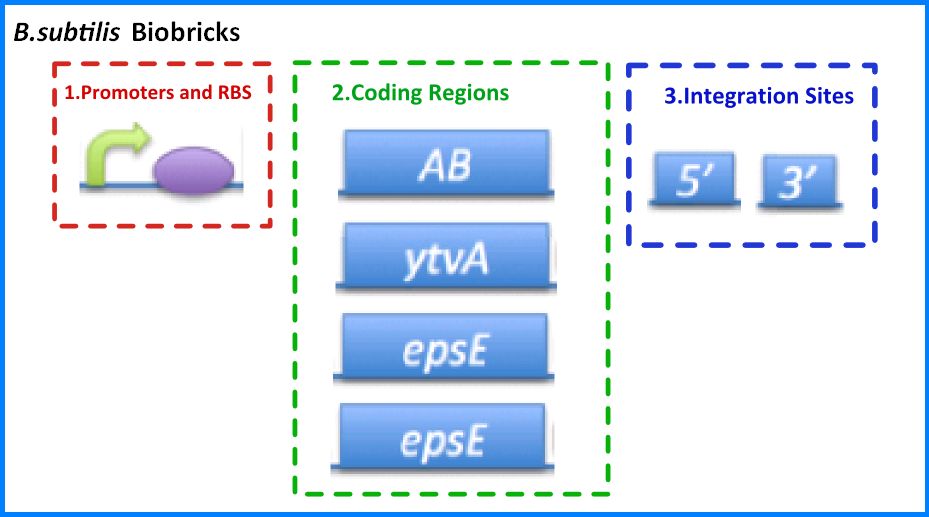
| - | {{Imperial/Box1|''B. subtilis'' | + | {{Imperial/Box1|''B. subtilis'' BioBricks|In order to pursue our project we had to design and build a variety of BioBrick parts for ''B. subtilis''. These can be split up into three groups: |
*'''Promoter and Ribosome Binding Sites (RBS)''' - These promoter and RBS combinations include a number of inducible and constitutive promoters combined with weak and strong ribosome binding sites. | *'''Promoter and Ribosome Binding Sites (RBS)''' - These promoter and RBS combinations include a number of inducible and constitutive promoters combined with weak and strong ribosome binding sites. | ||
*'''Coding Regions''' - These parts include specific coding regions concerned with our project such as biomaterials, and more general coding regions such as antibiotic resistance genes that can be used for selection markers. | *'''Coding Regions''' - These parts include specific coding regions concerned with our project such as biomaterials, and more general coding regions such as antibiotic resistance genes that can be used for selection markers. | ||
Revision as of 21:26, 29 October 2008
|
|||||||||||||||||||
 "
"