Team:Prairie View/Notebook
From 2008.igem.org
RAUL CUERO (Talk | contribs) |
RAUL CUERO (Talk | contribs) |
||
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Image:Prairie_View_iGEM.jpg|700px|center|]] | ||
| + | |||
{| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | |||
!align="center"|[[Team:Prairie_View|Home]] | !align="center"|[[Team:Prairie_View|Home]] | ||
!align="center"|[[Team:Prairie_View/Team|The Team]] | !align="center"|[[Team:Prairie_View/Team|The Team]] | ||
| Line 6: | Line 9: | ||
!align="center"|[[Team:Prairie_View/Modeling|Modeling]] | !align="center"|[[Team:Prairie_View/Modeling|Modeling]] | ||
!align="center"|[[Team:Prairie_View/Notebook|Notebook]] | !align="center"|[[Team:Prairie_View/Notebook|Notebook]] | ||
| - | |} | + | |}<br> |
| - | + | <h1>[[PV Protocols]]</h1> | |
| - | + | ||
| - | < | + | |
| - | + | ||
<h2>July</h2>Planning | <h2>July</h2>Planning | ||
| - | We began our project by discussing the | + | We began our project by discussing the interaction and function of an Electronic Nose (E-Nose) for assessing, interpreting and analyzing the parameters of data results. <br> |
| - | ENOSE FUNCTION | + | ENOSE FUNCTION<br> |
| - | Through planning out our laboratory project, we decided that designing our parts in a way that they can be assembled in a modular fashion could best be achieved by | + | Through planning out our laboratory project, we decided that designing our parts in a way that they can be assembled in a modular fashion could best be achieved by having compatible restriction enzymes site to flank each gene. We then had to research the process of PCR to get a better understanding of how to design primers for this approach, compiled laboratory techniques that we would be using during the project, and consulted the literature for ideas on design and assembly. <br> |
| + | We designed primers for each part that was synthesized for amplification in PCR. An oligo was designed containing the RBS sequence and was surrounded with compatible restriction sites which would be ligated in front of each part.<br> | ||
| + | |||
| + | <h2>August</h2>Primer Testing, PCR and Digestion | ||
| + | The first attempts at running our primers in PCR were more or less unsuccessful, only amplifying one or two of our parts. <br> | ||
| + | |||
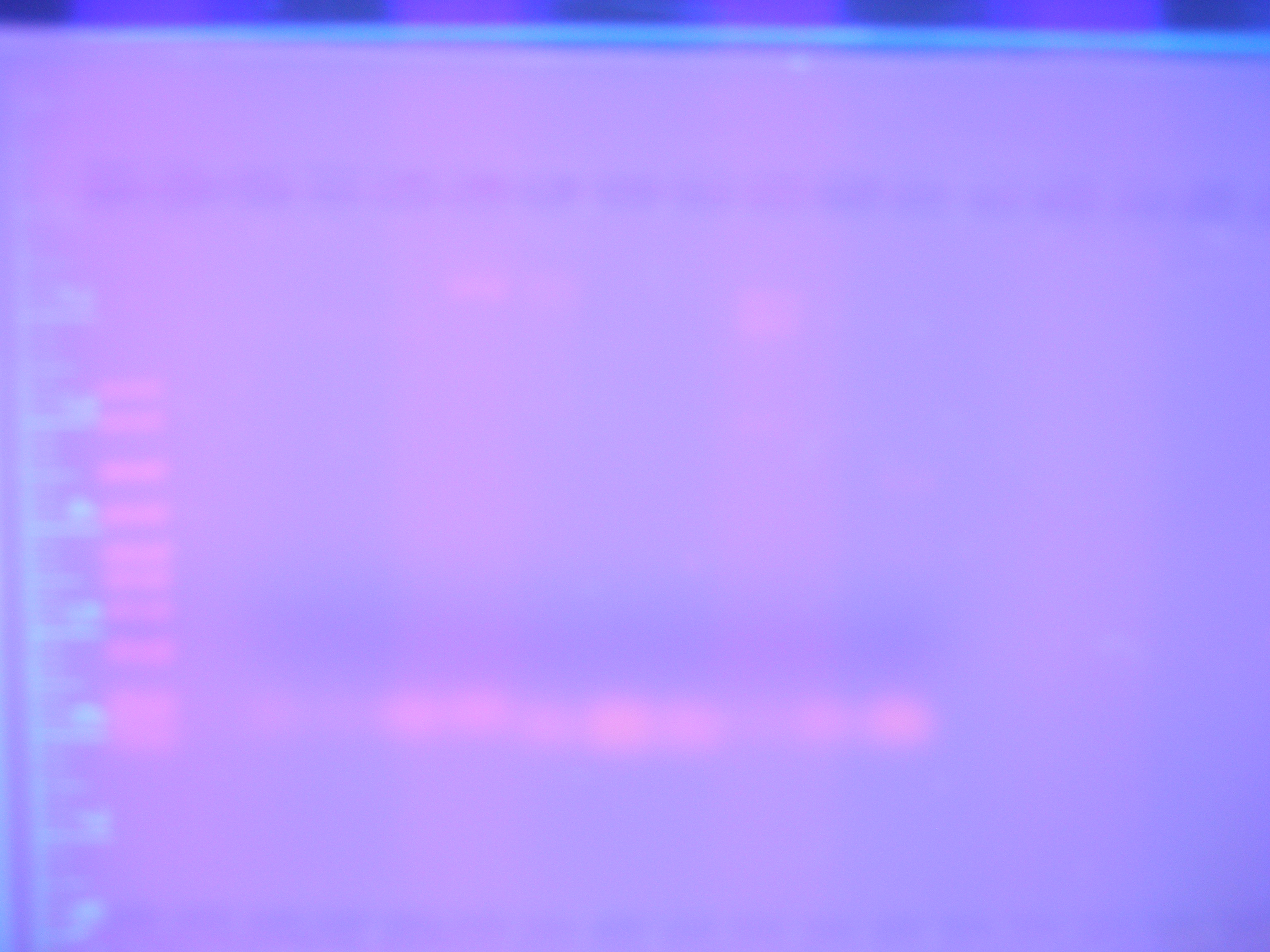
| + | |[[Image:Figure3.jpg|400px|center|]]<br> | ||
| + | |||
| + | Figure 1 PCR 8/05/08<br> | ||
| + | (Marker, Cytochrome, Vanadium, Calcium, FhuA, Ton B, Nickel B, Nickel A) | ||
| + | <br><br> | ||
| + | |||
| + | We spent a fair amount of time adjusting the parameters of the PCR program that we ran in order to optimize the amplification of our parts using the primers that we had designed. Roughly half of our primer was annealing to the gene and the other half was an added tail containing restriction sites. <br><br> | ||
| + | After running PCR with our designed primers on each part, we digested the parts that were successfully amplified to be ligated behind an RBS and into our expression vector. Performing this two step ligation connecting each part to a RBS proved complicating. Through multiple attempts, we tried varying the concentrations and volumes for the ligation but would see few colonies on media plates. | ||
| + | |||
| - | |||
| - | |||
| - | |||
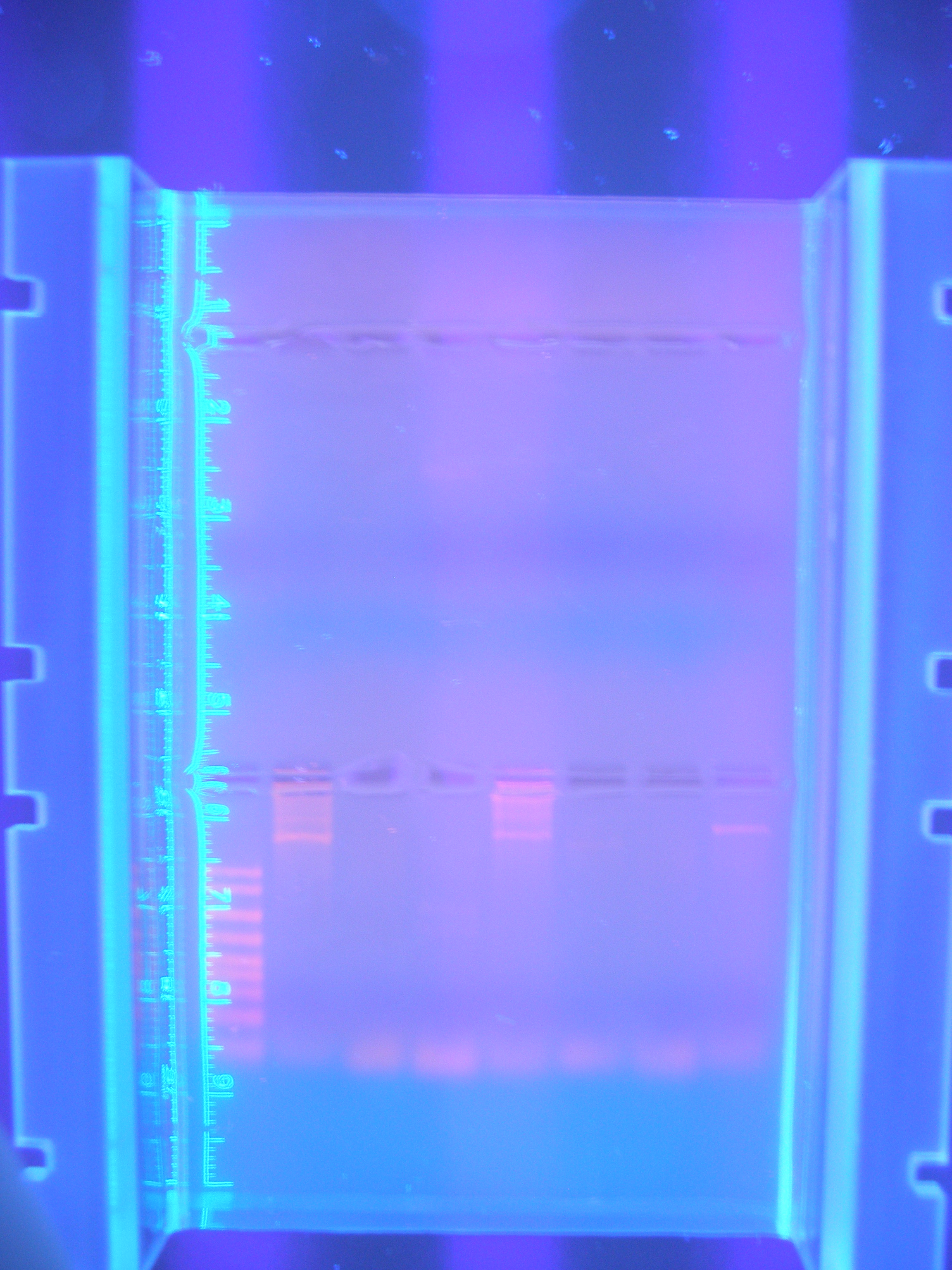
| - | + | |[[Image:Figure2.jpg|250px|center|]]<br> | |
| - | + | ||
| - | <br> | + | |
| - | Figure 2 Parts1 (9.25.08) | + | Figure 2 Parts1 (9.25.08)<br> |
This gel shows our PCR digested. | This gel shows our PCR digested. | ||
| - | + | Cytochrome C: 354 - | |
| - | + | Calcium: 756 - | |
| - | + | Ton B: 768 - | |
| - | + | Vanadium: 1254 - | |
| - | + | Nickel A: 1614 - | |
| - | + | Nickel B: 978 - | |
| - | + | FhuA: 2277 - | |
| - | + | Marker - | |
| - | + | ---- | |
| - | + | ||
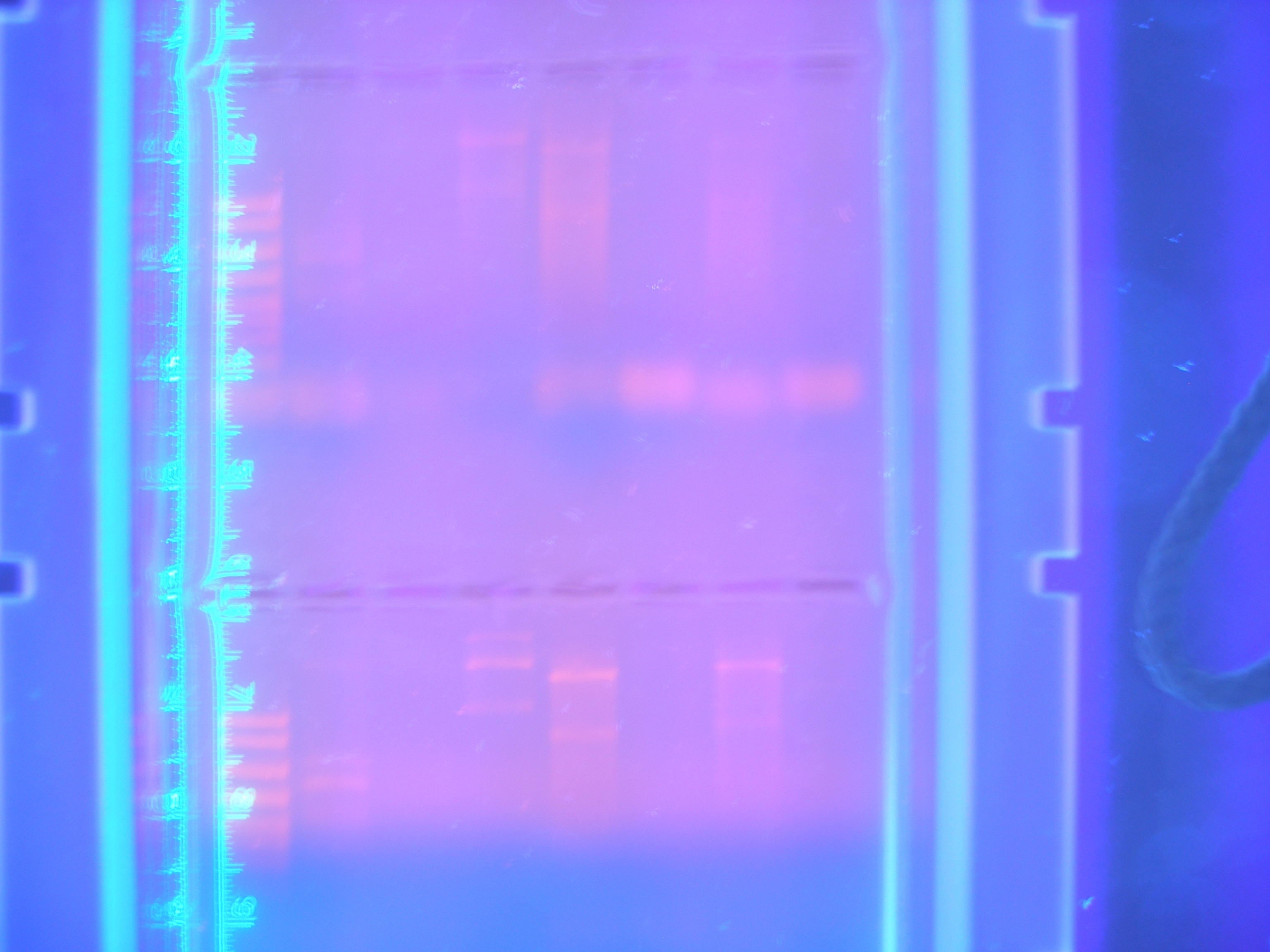
| + | [[Image:Figure1.jpg|250px|center|]] <br> | ||
| + | Figure 3 DigPCRparts(8.25.08) | ||
| - | |||
| - | |||
| - | + | Nickle A, Cytochrome C, and Ton B, And FhuA were the only parts that showed consistenly. FhuA and Nik.A would show bands at correct sizes, however the bands for Cytochrome C and Ton B were the wrong sizes. This meant to us that there was still a problem with the PCR reaction. It could have either been that the primers that were designed were not working in the way that we had hoped or that there was a parameter of the PCR program that was not allowing amplification of each part. | |
| + | <h2>September</h2> Redesigning of primers and Assembly/Digesting and Ligation. | ||
| - | + | To eliminate complications of ligating parts to a RBS, we designed new primers that included the RBS sequence into the PCR product. By doing this, we avoided the two-step ligation process. Some of the genes that we were going to include into our ligations contained the same restriction enzyme sites internally that would be to assemble the parts together. To avoid digesting our gene in the assembly process, one point mutations had to be introduced that would change one base of the restriction site while maintaining the same translated amino acid. To do this, mutation primers were designed and run on PCR to introduce the point mutations. | |
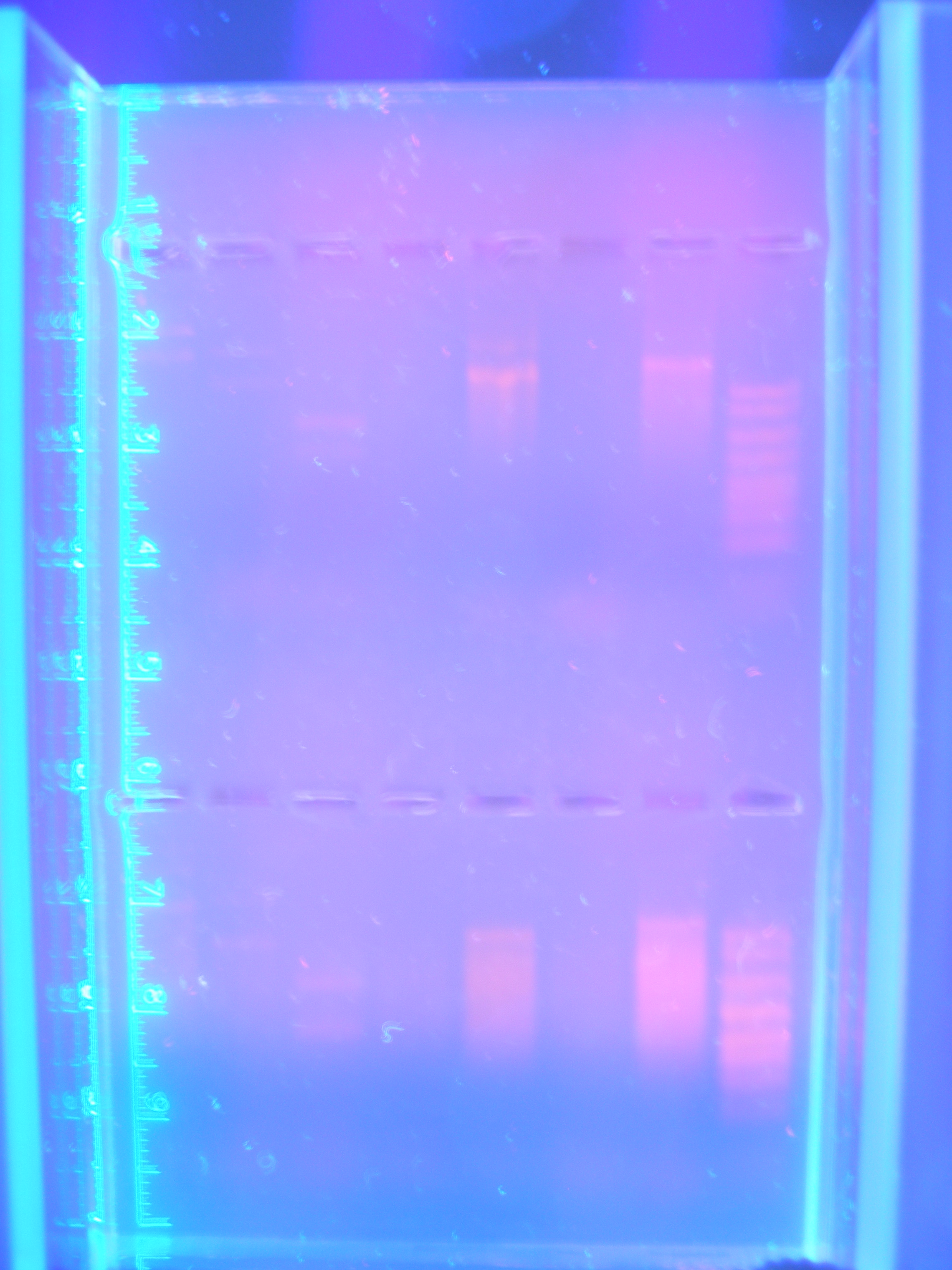
| - | + | [[Image:Figure4.jpg|250px|center|]]<br> | |
| - | + | Figure 4: Parts PCR with New Primers (9.11.08)<br><br> | |
| - | + | ---- | |
| - | Figure | + | |
| + | |||
| + | [[Image:Figure5.jpg|250px|center|]]<br> | ||
| + | Figure 5: Digested Parts (9.16.08)<br><br> | ||
| + | NA1, NA2, NA3, FhuA(long, EcFhuA, EN2 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[Image:Figure6.jpg|250px|center|]]<br> | ||
| + | Figure 6: PCR Mutation (9.28.08)<br><br> | ||
| + | For this particular mutation we used upstreams and downstreams: | ||
| + | VIP(up), VIP(dwn), MgTu, MgTd, MgBu, MgBd, Znu, Znd:<br> | ||
| + | VIP(FR), VIP(R), MgTu(FR), MgTd(R), MgBu(FR), MgBd(R), Znu(FR), Znd(R) | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[Image:Figure7.jpg|250px|center|]]<br> | ||
| + | Figure 7: PCR Mutation (9.30.08)<br><br> | ||
| + | |||
| + | Fig 7 shows mutations of: | ||
| + | Marker,pET, VIP, MgT, MgB, Zn, Marker | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<h2>October</h2>- Testing Ligations and inputting data for E-Nose Analysis | <h2>October</h2>- Testing Ligations and inputting data for E-Nose Analysis | ||
| - | We tested our ligations in vivo. The testing was composed of growing up ligation expressing cultures, taking samples at 0,12,18,24,36, and 48-hours and extracting their plasmids. Measurements were taken of OD, DNA concentration, DNA fluorescence and ATP concentration to determine the effects of the ligations within the cell. | + | We tested our ligations in vivo. The testing was composed of growing up ligation expressing cultures, taking cultural samples at 0,12,18,24,36, and 48-hours and extracting their plasmids. Measurements were taken of OD, DNA concentration, DNA fluorescence and ATP concentration to determine the effects of the ligations within the cell. |
DATA | DATA | ||
Latest revision as of 23:31, 29 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Contents |
PV Protocols
July
PlanningWe began our project by discussing the interaction and function of an Electronic Nose (E-Nose) for assessing, interpreting and analyzing the parameters of data results.
ENOSE FUNCTION
Through planning out our laboratory project, we decided that designing our parts in a way that they can be assembled in a modular fashion could best be achieved by having compatible restriction enzymes site to flank each gene. We then had to research the process of PCR to get a better understanding of how to design primers for this approach, compiled laboratory techniques that we would be using during the project, and consulted the literature for ideas on design and assembly.
We designed primers for each part that was synthesized for amplification in PCR. An oligo was designed containing the RBS sequence and was surrounded with compatible restriction sites which would be ligated in front of each part.
August
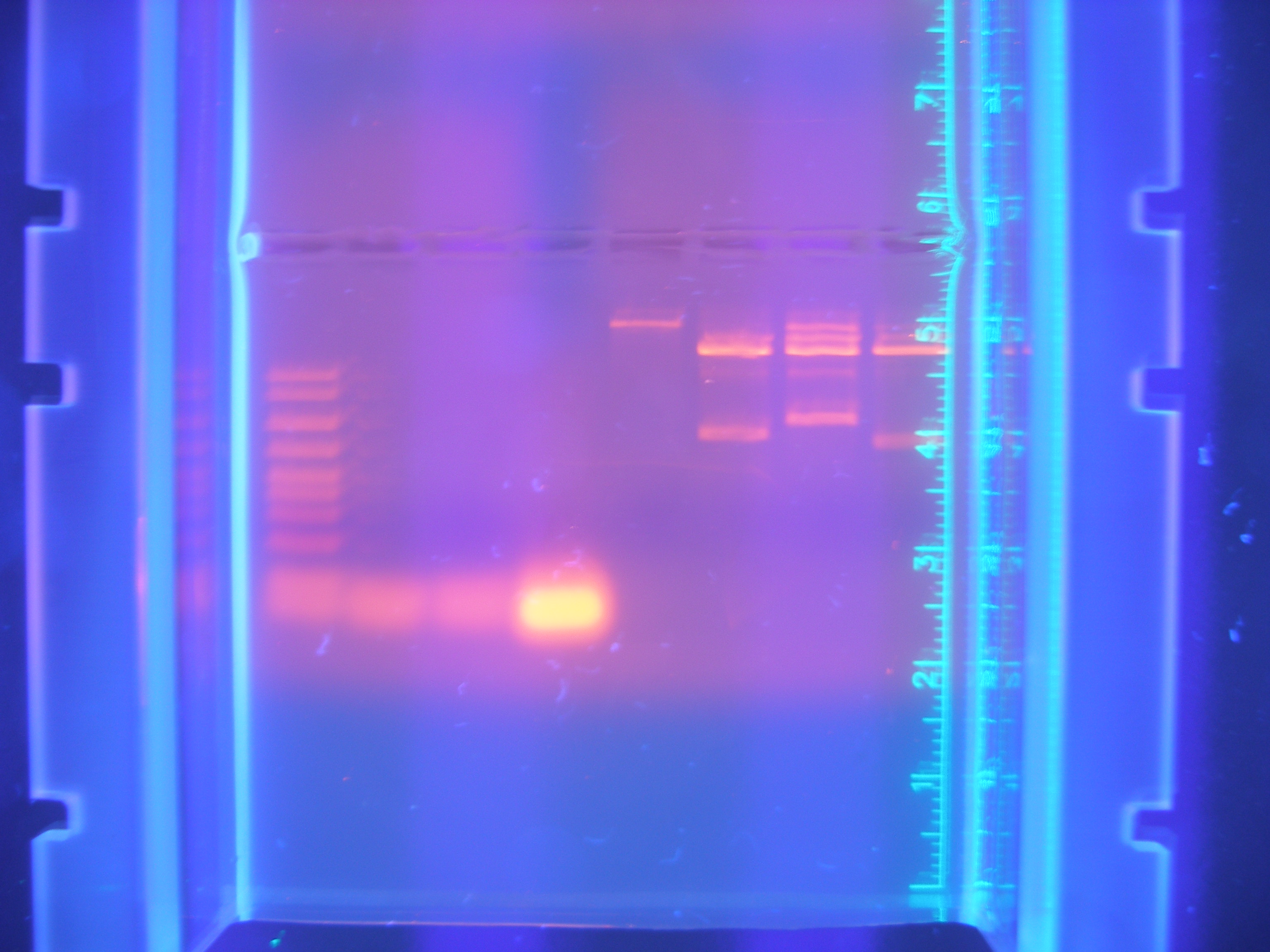
Primer Testing, PCR and DigestionThe first attempts at running our primers in PCR were more or less unsuccessful, only amplifying one or two of our parts.
Figure 1 PCR 8/05/08
(Marker, Cytochrome, Vanadium, Calcium, FhuA, Ton B, Nickel B, Nickel A)
We spent a fair amount of time adjusting the parameters of the PCR program that we ran in order to optimize the amplification of our parts using the primers that we had designed. Roughly half of our primer was annealing to the gene and the other half was an added tail containing restriction sites.
After running PCR with our designed primers on each part, we digested the parts that were successfully amplified to be ligated behind an RBS and into our expression vector. Performing this two step ligation connecting each part to a RBS proved complicating. Through multiple attempts, we tried varying the concentrations and volumes for the ligation but would see few colonies on media plates.
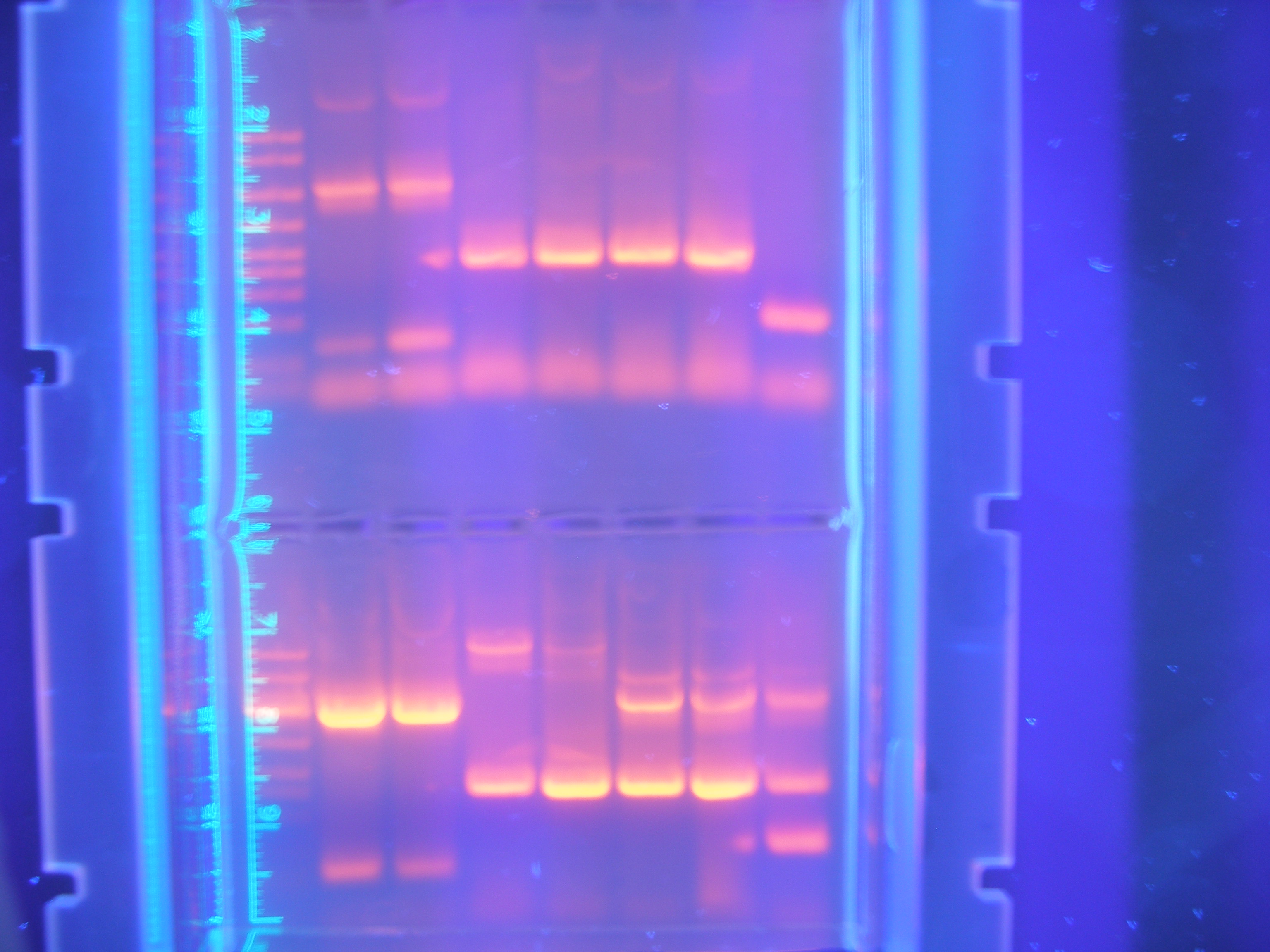
Figure 2 Parts1 (9.25.08)
This gel shows our PCR digested.
Cytochrome C: 354 -
Calcium: 756 -
Ton B: 768 -
Vanadium: 1254 -
Nickel A: 1614 -
Nickel B: 978 -
FhuA: 2277 -
Marker -
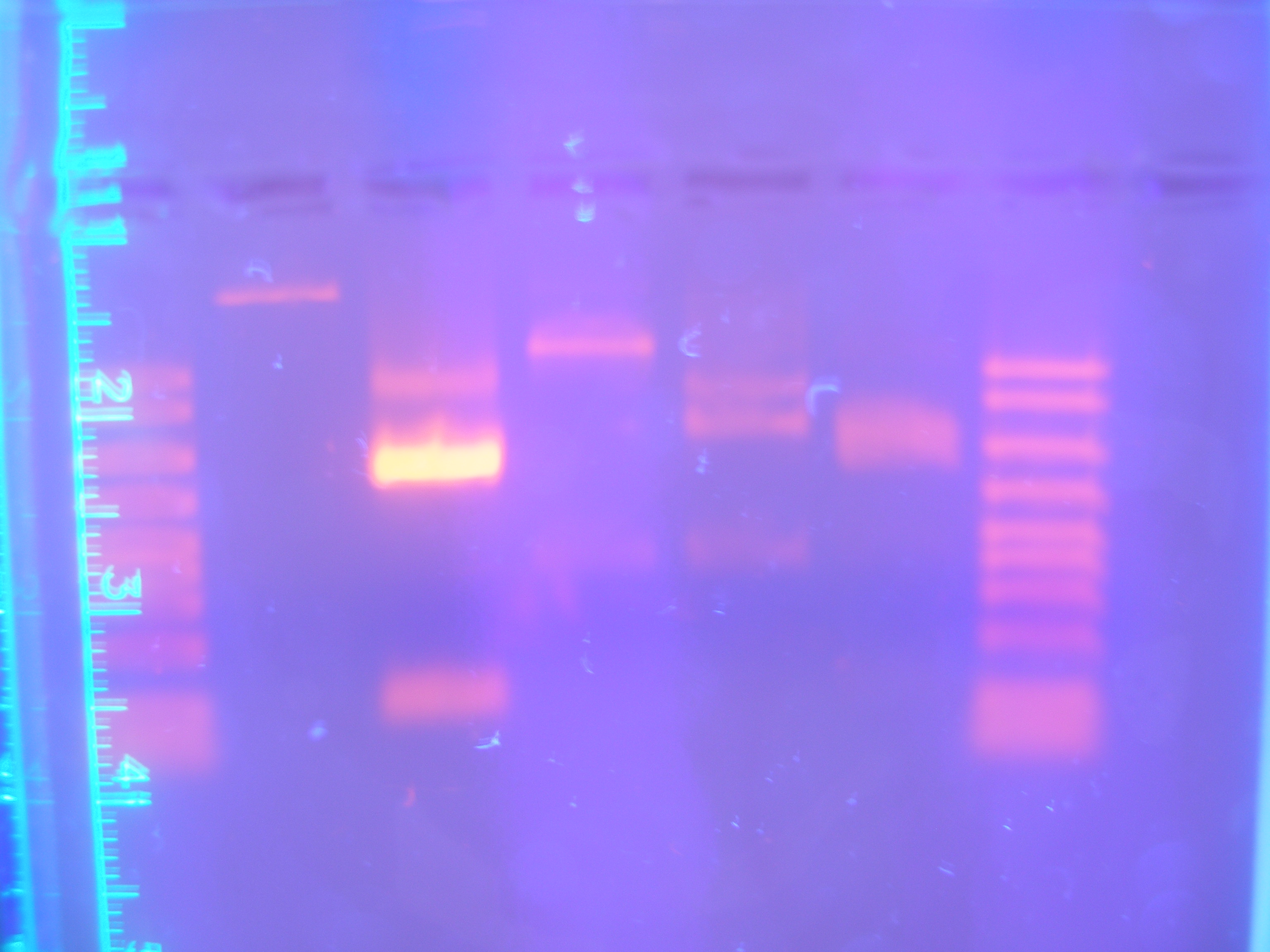
Figure 3 DigPCRparts(8.25.08)
Nickle A, Cytochrome C, and Ton B, And FhuA were the only parts that showed consistenly. FhuA and Nik.A would show bands at correct sizes, however the bands for Cytochrome C and Ton B were the wrong sizes. This meant to us that there was still a problem with the PCR reaction. It could have either been that the primers that were designed were not working in the way that we had hoped or that there was a parameter of the PCR program that was not allowing amplification of each part.
September
Redesigning of primers and Assembly/Digesting and Ligation.
To eliminate complications of ligating parts to a RBS, we designed new primers that included the RBS sequence into the PCR product. By doing this, we avoided the two-step ligation process. Some of the genes that we were going to include into our ligations contained the same restriction enzyme sites internally that would be to assemble the parts together. To avoid digesting our gene in the assembly process, one point mutations had to be introduced that would change one base of the restriction site while maintaining the same translated amino acid. To do this, mutation primers were designed and run on PCR to introduce the point mutations.
Figure 4: Parts PCR with New Primers (9.11.08)
Figure 5: Digested Parts (9.16.08)
NA1, NA2, NA3, FhuA(long, EcFhuA, EN2
Figure 6: PCR Mutation (9.28.08)
For this particular mutation we used upstreams and downstreams:
VIP(up), VIP(dwn), MgTu, MgTd, MgBu, MgBd, Znu, Znd:
VIP(FR), VIP(R), MgTu(FR), MgTd(R), MgBu(FR), MgBd(R), Znu(FR), Znd(R)
Figure 7: PCR Mutation (9.30.08)
Fig 7 shows mutations of: Marker,pET, VIP, MgT, MgB, Zn, Marker
October
- Testing Ligations and inputting data for E-Nose AnalysisWe tested our ligations in vivo. The testing was composed of growing up ligation expressing cultures, taking cultural samples at 0,12,18,24,36, and 48-hours and extracting their plasmids. Measurements were taken of OD, DNA concentration, DNA fluorescence and ATP concentration to determine the effects of the ligations within the cell. DATA
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"