Team:NYMU-Taipei/Project/Time Regulation/Cyanoxilator
From 2008.igem.org
Blackrabbit (Talk | contribs) |
Blackrabbit (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Header}} | {{:Team:NYMU-Taipei/Header}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | *Nakajima, M., K. Imai, et al. (2005). "Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro." Science 308(5720): 414-5. | + | == Motivation == |
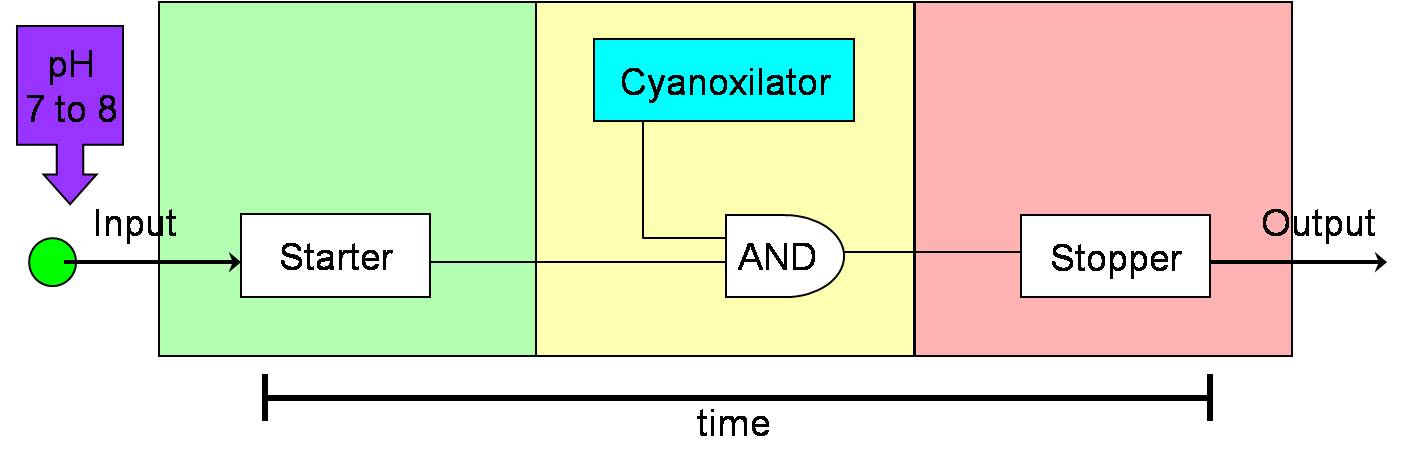
| - | + | [[Image:NYMU Timer cyano.jpg|600px|right]] | |
| - | + | * To enhance the the effect of the micro dialysis machine in the intestine lumen, the half life of the micro dialysis machine is designed to be prolonged. However, since our micro dialysis machine is made in the chassis of ''E. coli'', there are two possible risks while staying in the intestine too long. One is that the ''E. coli'' may be apoptosis or necrosis due to large amount of the metabolite wastes it absorbed. The other is the bacteria balance in the intestine may be disturbed thanks to the ''E. coli'' being added. Thus, it's crucial and necessary to have a timer in our micro dialysis machine. | |
| + | * Besides, the timer has many biological and engineering application, such as artificial pacemakers, biological clocks, etc. | ||
| + | |||
| + | == Literature review == | ||
| + | ===Overview of the cyanobacterial Kai oscillators [3]=== | ||
| + | [[Image:NYMU Cyanobacteria.JPG|thumb|[http://www.dkimages.com/discover/DKIMAGES/Discover/Home/Plants/Fungi-Monera-Protista/Cyanobacteria/Cyanobacteria-2.html Cyanobacteria]]] | ||
| + | Kai proteins globally regulate circadian gene expression of cyanobacteria. The | ||
| + | KaiC phosphorylation cycle, which persists even without transcription or | ||
| + | translation, is assumed to be a basic timing process of the circadian clock. The self-sustainable oscillation of KaiC phosphorylation had been reconstituted in | ||
| + | vitro by incubating KaiC with KaiA, KaiB, and adenosine triphosphate. The period | ||
| + | of the in vitro oscillation was stable despite temperature change (temperature | ||
| + | compensation), and the circadian periods observed in vivo in KaiC mutant | ||
| + | strains were consistent with those measured in vitro. Therefore, it's of great possibility that the cyanobacterial Kai oscillators can also be reconstituted in ''E. coli.'' | ||
| + | |||
| + | ===The relationship between KaiABC proteins and their output pathways [1]=== | ||
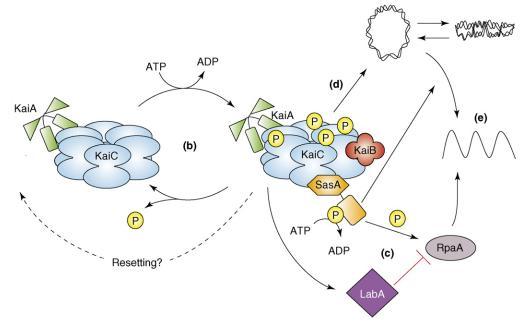
| + | [[Image:NYMU KaiABC.JPG|center]] | ||
| + | * (B) The oscillation in KaiC phosphorylation results from the opposing actions of KaiA, which stimulates KaiC autokinase activity, and KaiB, which attenuates the positive effect of KaiA. Increasing evidence supports the hypothesis that the phosphorylation state of KaiC affects the ability of the clock to reset properly. | ||
| + | * (C) The SasA protein interacts with KaiC through its N terminus (hexagonal domain), which is similar in sequence but not structure to KaiB. Temporal information is transduced from the Kai oscillator to SasA through the stimulation of SasA autophosphorylation by KaiC. RpaA is activated by SasA through transfer of a phosphoryl group. LabA acts as a mediator of negative regulation from KaiC and is predicted to repress RpaA function. | ||
| + | * (D) In addition, the Kai oscillator regulates the compaction rhythm of the cyanobacterial nucleoid (double-stranded loop), which would control accessibility of transcriptional machinery to promoter regions. | ||
| + | * (E) SasA is not required for rhythmic chromosome compaction, but it is necessary (as is RpaA) for overt rhythmicity of gene expression. Arrows and perpendicular lines indicate, respectively, positive and negative regulation. | ||
| + | |||
| + | == Aims == | ||
| + | # Building KaiABC proteins, SasA and RpaA in ''E. coli''. | ||
| + | # Measuring the output of the phosphorylation status of KaiC through pPhoB, pOmpC and pOmpF. | ||
| + | # Tuning the period of oscillation by regulating the amount of KaiA. | ||
| + | |||
| + | == System Design == | ||
| + | === Aim 1: Building KaiABC proteins, SasA and RpaA in E. coli.=== | ||
| + | {| border=1 | ||
| + | ! States of oscillation | ||
| + | ! Status | ||
| + | |- | ||
| + | |[[Image:NYMU Design p.jpg|400px]] | ||
| + | | | ||
| + | *Interaction of C-KaiA with KaiC stimulates the autokinase activity of KaiC to result in a hyperphosphorylated KaiC protein. | ||
| + | *Temporal information is transduced from the Kai oscillator to RpaA through SasA. | ||
| + | |||
| + | |- | ||
| + | |[[Image:NYMU Design unp.jpg|400px]] | ||
| + | | | ||
| + | *KaiB interacts with phosphorylated KaiC, thereby decreasing the positive role of KaiA on KaiC and promoting dephosphorylation of KaiC. | ||
| + | *Non-phosphorylated KaiC is bound by KaiA and the cycle begins again. | ||
| + | |} | ||
| + | |||
| + | |||
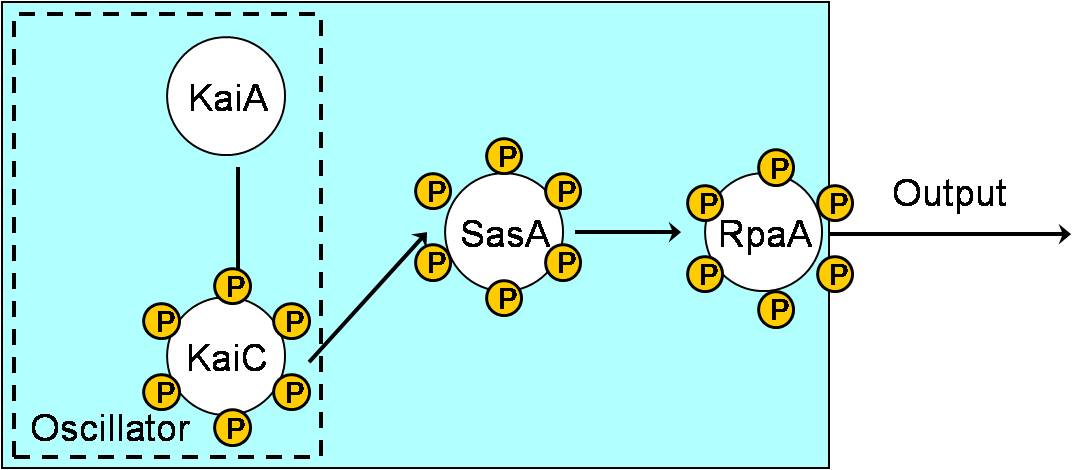
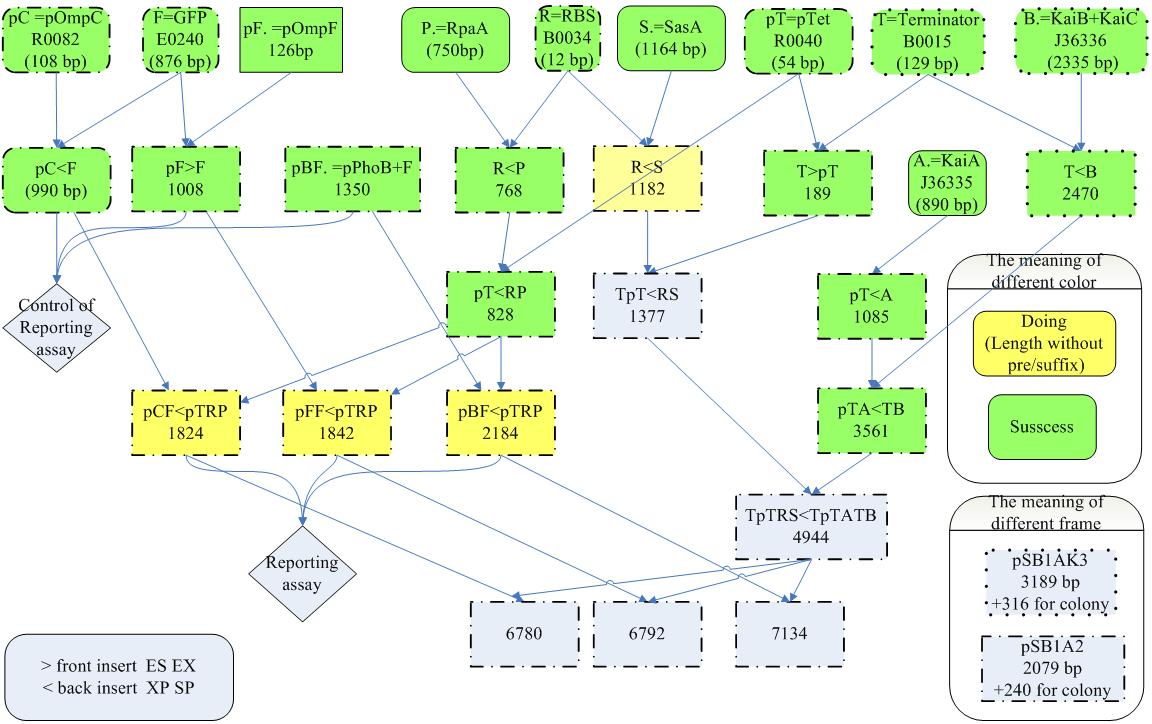
| + | ===Aim 2: Measuring the output of the phosphorylation status of KaiC through pPhoB, pOmpC and pOmpF. === | ||
| + | {| | ||
| + | |[[Image:NYMU Cyano reporting circuit.jpg|400px|left]] | ||
| + | | | ||
| + | * Although the direct target of RpaA is still unknown, there still some predicted targets. | ||
| + | * Since SasA and RpaA is highly homologous to EnvZ and OmpR, respectively, we predicted that RpaA may also bind to the targets of OmpR. | ||
| + | * Besides,PhoB and OmpR are response regulators in the same subfamily and are highly homologous. Thus. PhoB may be a predicted target. Response regulators is composed of an N-terminal phosphorylation domain and a C-terminal DNA binding effector domain connected by a flexible interdomain linker. | ||
| + | |||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ===Aim 3: Tuning the period of oscillation by regulating the amount of KaiA. === | ||
| + | [[Image:NYMU KaiABCcircuit.JPG]] | ||
| + | * KaiA and KaiBC proteins is under the control of the inducible promoter in order to tune the period of oscillation by regulating the amount of KaiA | ||
| + | |||
| + | == Materials == | ||
| + | * [http://www.bio.tamu.edu/synecho/ ''S. elongatus'' PCC 7942] from [http://www.bio.tamu.edu/facmenu/faculty/GoldenS.htm Susan Golden] and Takao Kondo | ||
| + | * [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=Search&TermToSearch=txid1140 Genome sequences of PCC 7942 at NCBI] | ||
| + | |||
| + | == Method == | ||
| + | ===assembly process=== | ||
| + | [[Image:NYMU Cyanoassembly0923.jpg|700px]] | ||
| + | === Extract Cyanobacteria Genomic DNA === | ||
| + | * [http://www.protocol-online.org/prot/Protocols/Rapid-DNA-Extraction-from-Cyanobacteria-3993.html Rapid DNA Extraction from Cyanobacteria] | ||
| + | <pre> | ||
| + | Materials | ||
| + | |||
| + | * Phenol | ||
| + | * Chloroform | ||
| + | * Absolute ethanol | ||
| + | * 70% ethanol | ||
| + | * TE buffer (10T/1E), pH 8.0 | ||
| + | * 10% SDS | ||
| + | * 50 mg/ml lysozyme | ||
| + | * 5 M NaCl | ||
| + | |||
| + | Procedure | ||
| + | |||
| + | 1. Macerate Cyanobacteria in TE Buffer (10T/1E) | ||
| + | 2. Pellet the cells by centrifugation for 2 min at 10,000 rpm at room temperature | ||
| + | 3. Remove supernatant. Add 500 µl TE buffer (10T/1E). | ||
| + | 4. Add 1% SDS and 50 µl of 50 mg/ml lysozyme stock solution | ||
| + | 5. Keep at 70°C for 15 min. | ||
| + | 6. Add equal volume of Phenol : Chloroform | ||
| + | 7. Centrifuge for 10 min at 10,000 rpm at room temperature | ||
| + | 8. Take supernatant and extract it twice using 100% Chloroform extraction procedure. | ||
| + | 9. Finally take supernatant and add 0.1 volume of 5M NaCl and 2 volumes of 100% ethanol and keep for precipitation at –20°C for 2 hrs | ||
| + | 10. Centrifuge the tubes for 30 min at 15,000 rpm at 10°C | ||
| + | 11. Wash the pellet twice by 70% ethanol by centrifuging it at 10,000 rpm for 10 minutes. | ||
| + | 12. Dry the pellet and resuspend in TE buffer (10T/1E) | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | |||
| + | == [[Team:NYMU-Taipei/Project/Time_Regulation/Results_of_Cyanoxilator|Results]]== | ||
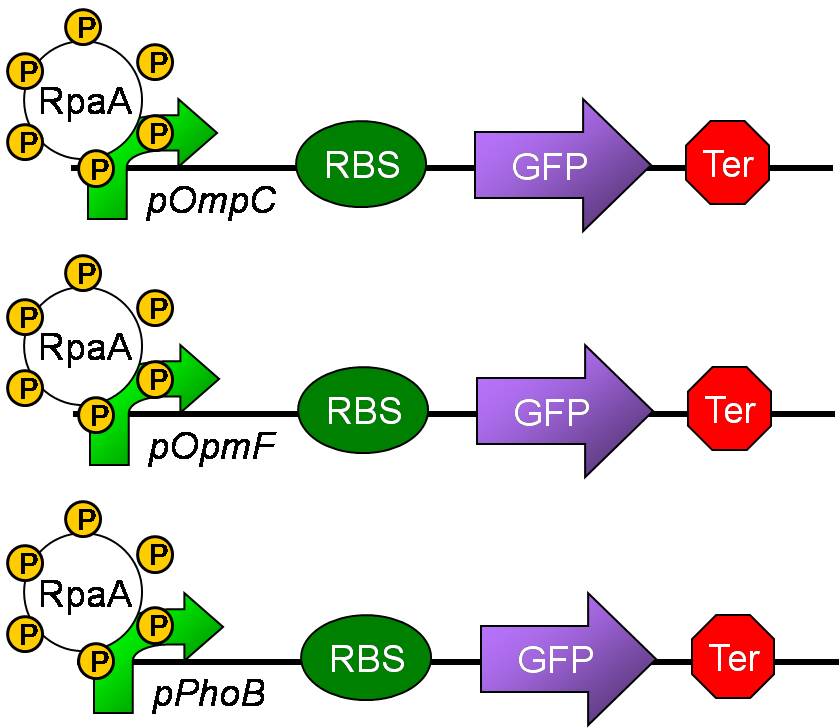
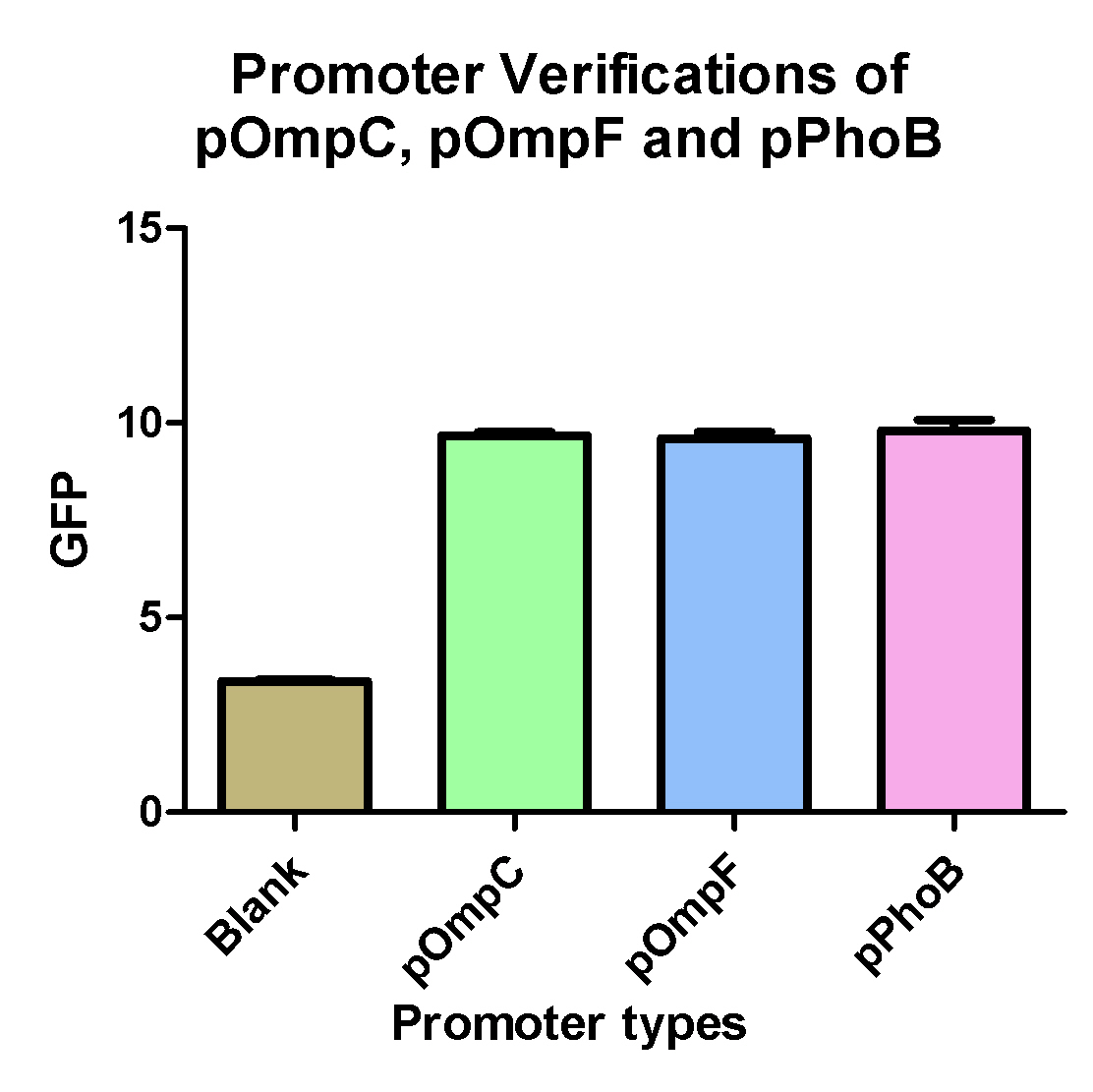
| + | ===Promoter Verifications of pOmpC, pOmpF and pPhoB=== | ||
| + | {| | ||
| + | | | ||
| + | [[Image:NYMU Cyano promoter reporter.jpg|350px]] | ||
| + | | | ||
| + | * pOmpC(BBa_R0082) are an already exist biobrick but so far without any verifications | ||
| + | * pOmpF and pPhoB are two biobricks we created from PCR of ''E. coli'' K12. | ||
| + | * Experimental results shows that the above three promoters can promote the expression of the GFP compare to blank which is GFp only without any promoter. | ||
| + | |- | ||
| + | |} | ||
| + | * For other experimental data, please click [[Team:NYMU-Taipei/Project/Time_Regulation/Results_of_Cyanoxilator|here]] | ||
| + | |||
| + | == Reference == | ||
| + | # Mackey, S. R. and S. S. Golden (2007). "Winding up the cyanobacterial circadian clock." Trends in Microbiology 15(9): 381-8. | ||
| + | # Y. Murayama, T. Oyama, T. Kondo, J Bacteriol 190, 1691 (Mar. 2008). | ||
| + | # Nakajima, M., K. Imai, et al. (2005). "Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro." Science 308(5720): 414-5. | ||
| + | # Takai, N., M. Nakajima, et al. (2006). "A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria.[see comment]." Proceedings of the National Academy of Sciences of the United States of America 103(32): 12109-14. | ||
| + | # [http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria iGEM 2006 Harvard U.] | ||
| + | # S. Kutsuna et al., J Bacteriol 189, 7690 (Nov, 2007). | ||
| + | |||
| + | == Acknowledgments == | ||
| + | [[Image:NYMU Takao.jpg]][[Image:NYMU Susan.jpg]] | ||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 00:25, 30 October 2008
| Home | Project Overview: | pH Sensor | Attachment | Time Regulation | Waste Removal | Experiments and Parts | About Us |
Contents |
Motivation
- To enhance the the effect of the micro dialysis machine in the intestine lumen, the half life of the micro dialysis machine is designed to be prolonged. However, since our micro dialysis machine is made in the chassis of E. coli, there are two possible risks while staying in the intestine too long. One is that the E. coli may be apoptosis or necrosis due to large amount of the metabolite wastes it absorbed. The other is the bacteria balance in the intestine may be disturbed thanks to the E. coli being added. Thus, it's crucial and necessary to have a timer in our micro dialysis machine.
- Besides, the timer has many biological and engineering application, such as artificial pacemakers, biological clocks, etc.
Literature review
Overview of the cyanobacterial Kai oscillators [3]
Kai proteins globally regulate circadian gene expression of cyanobacteria. The KaiC phosphorylation cycle, which persists even without transcription or translation, is assumed to be a basic timing process of the circadian clock. The self-sustainable oscillation of KaiC phosphorylation had been reconstituted in vitro by incubating KaiC with KaiA, KaiB, and adenosine triphosphate. The period of the in vitro oscillation was stable despite temperature change (temperature compensation), and the circadian periods observed in vivo in KaiC mutant strains were consistent with those measured in vitro. Therefore, it's of great possibility that the cyanobacterial Kai oscillators can also be reconstituted in E. coli.
The relationship between KaiABC proteins and their output pathways [1]
- (B) The oscillation in KaiC phosphorylation results from the opposing actions of KaiA, which stimulates KaiC autokinase activity, and KaiB, which attenuates the positive effect of KaiA. Increasing evidence supports the hypothesis that the phosphorylation state of KaiC affects the ability of the clock to reset properly.
- (C) The SasA protein interacts with KaiC through its N terminus (hexagonal domain), which is similar in sequence but not structure to KaiB. Temporal information is transduced from the Kai oscillator to SasA through the stimulation of SasA autophosphorylation by KaiC. RpaA is activated by SasA through transfer of a phosphoryl group. LabA acts as a mediator of negative regulation from KaiC and is predicted to repress RpaA function.
- (D) In addition, the Kai oscillator regulates the compaction rhythm of the cyanobacterial nucleoid (double-stranded loop), which would control accessibility of transcriptional machinery to promoter regions.
- (E) SasA is not required for rhythmic chromosome compaction, but it is necessary (as is RpaA) for overt rhythmicity of gene expression. Arrows and perpendicular lines indicate, respectively, positive and negative regulation.
Aims
- Building KaiABC proteins, SasA and RpaA in E. coli.
- Measuring the output of the phosphorylation status of KaiC through pPhoB, pOmpC and pOmpF.
- Tuning the period of oscillation by regulating the amount of KaiA.
System Design
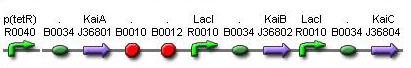
Aim 1: Building KaiABC proteins, SasA and RpaA in E. coli.
Aim 2: Measuring the output of the phosphorylation status of KaiC through pPhoB, pOmpC and pOmpF.
|
Aim 3: Tuning the period of oscillation by regulating the amount of KaiA.
- KaiA and KaiBC proteins is under the control of the inducible promoter in order to tune the period of oscillation by regulating the amount of KaiA
Materials
- [http://www.bio.tamu.edu/synecho/ S. elongatus PCC 7942] from [http://www.bio.tamu.edu/facmenu/faculty/GoldenS.htm Susan Golden] and Takao Kondo
- [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=Search&TermToSearch=txid1140 Genome sequences of PCC 7942 at NCBI]
Method
assembly process
Extract Cyanobacteria Genomic DNA
- [http://www.protocol-online.org/prot/Protocols/Rapid-DNA-Extraction-from-Cyanobacteria-3993.html Rapid DNA Extraction from Cyanobacteria]
Materials
* Phenol
* Chloroform
* Absolute ethanol
* 70% ethanol
* TE buffer (10T/1E), pH 8.0
* 10% SDS
* 50 mg/ml lysozyme
* 5 M NaCl
Procedure
1. Macerate Cyanobacteria in TE Buffer (10T/1E)
2. Pellet the cells by centrifugation for 2 min at 10,000 rpm at room temperature
3. Remove supernatant. Add 500 µl TE buffer (10T/1E).
4. Add 1% SDS and 50 µl of 50 mg/ml lysozyme stock solution
5. Keep at 70°C for 15 min.
6. Add equal volume of Phenol : Chloroform
7. Centrifuge for 10 min at 10,000 rpm at room temperature
8. Take supernatant and extract it twice using 100% Chloroform extraction procedure.
9. Finally take supernatant and add 0.1 volume of 5M NaCl and 2 volumes of 100% ethanol and keep for precipitation at –20°C for 2 hrs
10. Centrifuge the tubes for 30 min at 15,000 rpm at 10°C
11. Wash the pellet twice by 70% ethanol by centrifuging it at 10,000 rpm for 10 minutes.
12. Dry the pellet and resuspend in TE buffer (10T/1E)
Results
Promoter Verifications of pOmpC, pOmpF and pPhoB
|
- For other experimental data, please click here
Reference
- Mackey, S. R. and S. S. Golden (2007). "Winding up the cyanobacterial circadian clock." Trends in Microbiology 15(9): 381-8.
- Y. Murayama, T. Oyama, T. Kondo, J Bacteriol 190, 1691 (Mar. 2008).
- Nakajima, M., K. Imai, et al. (2005). "Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro." Science 308(5720): 414-5.
- Takai, N., M. Nakajima, et al. (2006). "A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria.[see comment]." Proceedings of the National Academy of Sciences of the United States of America 103(32): 12109-14.
- [http://openwetware.org/wiki/IGEM:Harvard/2006/Cyanobacteria iGEM 2006 Harvard U.]
- S. Kutsuna et al., J Bacteriol 189, 7690 (Nov, 2007).
 "
"