Team:ETH Zurich/Wetlab/Overview
From 2008.igem.org
(→Outline) |
(→Outline) |
||
| Line 71: | Line 71: | ||
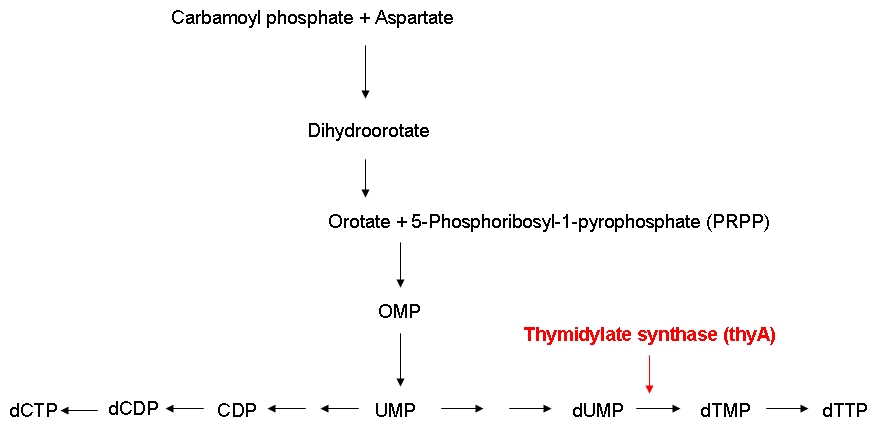
In order to be able to examine the growth behaviors of ''E. coli'' strains carrying differently sized genomes, we ordered an E. coli strain (MDS42) whose genome had been reduced by 15 % using a targeted deletion approach. Also, we ordered the wild-type strain which the MDS42 was derived from. For knocking out the thymidylate synthase, we decided to use phage transduction.<br><br> | In order to be able to examine the growth behaviors of ''E. coli'' strains carrying differently sized genomes, we ordered an E. coli strain (MDS42) whose genome had been reduced by 15 % using a targeted deletion approach. Also, we ordered the wild-type strain which the MDS42 was derived from. For knocking out the thymidylate synthase, we decided to use phage transduction.<br><br> | ||
'''Results:''' <br> | '''Results:''' <br> | ||
| - | We successfully knocked out the thymidylate synthase, both in the wild-type and the MDS42 ''E. coli'' strains. Also, we successfully performed growth experiments showing that the growth rate of thymidylate synthase knockout strains can be influenced by regulating the external thymidine supply. Additionally, we managed to label the wild-type and the MDS42 ''E. coli'' strains by transformation of low-copy plasmids encoding different reporter proteins. This enables us to keep track of the individual | + | We successfully knocked out the thymidylate synthase, both in the wild-type and the MDS42 ''E. coli'' strains. Also, we successfully performed growth experiments showing that the growth rate of thymidylate synthase knockout strains can be influenced by regulating the external thymidine supply. Additionally, we managed to label the wild-type and the MDS42 ''E. coli'' strains by transformation of low-copy plasmids encoding different reporter proteins. This enables us to keep track of the individual growth rates of the wild-type and MDS42 strain if grown in a mixed culture.<br><br> |
</div> | </div> | ||
|- | |- | ||
Revision as of 01:23, 30 October 2008
|
OverviewIn order to approach our goal of creating an E. coli strain carrying a minimal genome, there are three main problems that have to be overcome:
Chemostat selection: introduce a limitation that confers a growth advantage to organisms with smaller genomes Switch circuit: design a biobrick that provides for short-term synthesis of the desired gene products
Genome ReductionTo prove that in vivo restriction and religation is possible is fundamental to our project which relies on short-term expression of a restriction enzyme and a ligase. While the restriction enzyme will randomly cut DNA, the simultaneous or shortly delayed synthesis of the ligase should religate the DNA. If the DNA is cut at several sites, religation will lead to exclusion of chromosomal fragments in a random manner. Chemostat selectionIn the continuous culture of a chemostat, those organisms with the highest rate of proliferation will overgrow those with a smaller growth rate. In order to bypass the need of selecting for those E. coli which have successfully reduced their genomes by massive screening of thousands of clones, we need to introduce a constraint that confers a growth advantage to organisms with smaller genomes. We have chosen to introduce mutations in the nucleotide synthesis pathway to achieve this goal. This will render DNA replication the rate-limiting step of proliferation and therefore be advantageous to organisms with small genomes. Switch circuitExpression of restriction enzymes that cut genomic DNA inside the cell is likely to decrease viability. Actually, the Waterloo iGEM team is using restriction enzymes to kill the cell in their project this year. Therefore, construction of a switch circuit, which allows to restrict expression of the restriction enzyme to a short period of time, is a crucial part of the project.
Outline
|
 "
"