Team:Lethbridge CCS/Project
From 2008.igem.org
(→Method) |
(→7.) Transform & Use BioBrick) |
||
| (14 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
Our project centers on the development and optimization of a ligase-independent cloning (LIC) protocol. | Our project centers on the development and optimization of a ligase-independent cloning (LIC) protocol. | ||
| - | + | == '''Abstract''' == | |
''Ligase-Independent Cloning as a Standard for BioBrick Preparation'' | ''Ligase-Independent Cloning as a Standard for BioBrick Preparation'' | ||
| Line 9: | Line 9: | ||
drawback of several existing techniques is their dependence on ligase treatment, which is often problematic. We propose a ligase-independent cloning (LIC) method, based on the technique of [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)], as a possible standard for novel BioBrick preparation. Instead of short overhangs and ligase treatment, LIC uses long overhangs to circularize plasmid vectors for transformation without the use of ligase. The LIC method reduces the number of enzyme steps required for cloning, thus lending itself to easy adoption, automation, and real biological 'engineering.' | drawback of several existing techniques is their dependence on ligase treatment, which is often problematic. We propose a ligase-independent cloning (LIC) method, based on the technique of [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)], as a possible standard for novel BioBrick preparation. Instead of short overhangs and ligase treatment, LIC uses long overhangs to circularize plasmid vectors for transformation without the use of ligase. The LIC method reduces the number of enzyme steps required for cloning, thus lending itself to easy adoption, automation, and real biological 'engineering.' | ||
| - | + | == '''Rationale''' == | |
The underlying idea of iGEM is that an engineering approach can be applied to biology. Using a computer analogy, the goal is to have software (DNA sequences) that can be run on compatible hardware (microorganisms) anywhere in the world. | The underlying idea of iGEM is that an engineering approach can be applied to biology. Using a computer analogy, the goal is to have software (DNA sequences) that can be run on compatible hardware (microorganisms) anywhere in the world. | ||
| Line 27: | Line 27: | ||
Therefore, we chose to investigate ligase-independent cloning (LIC), a technique we believe can serve as a standard for BioBrick construction, in a way that is relatively easy to implement and that also lends itself to automation in the future. Our approach was based on a method described by [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)]. | Therefore, we chose to investigate ligase-independent cloning (LIC), a technique we believe can serve as a standard for BioBrick construction, in a way that is relatively easy to implement and that also lends itself to automation in the future. Our approach was based on a method described by [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)]. | ||
| - | + | == '''Method''' == | |
| - | 1.) Identify a | + | The intent is to be able to create a standard BioBrick from any DNA sequence of interest. |
| + | |||
| + | |||
| + | ==== 1.) Select Gene ==== | ||
| + | |||
| + | Identify a gene (DNA sequence) of interest | ||
{| | {| | ||
|width="2000px" align="center"|[[Image:gen_gene.png|General gene|400px]] | |width="2000px" align="center"|[[Image:gen_gene.png|General gene|400px]] | ||
|} | |} | ||
| - | 2.) Amplify, using PCR primers that contain the BioBrick prefix (brown) & suffix (purple) as well as a LIC overhang sequence (pink). | + | |
| + | ==== 2.) Amplify Gene ==== | ||
| + | |||
| + | Amplify, using PCR primers that contain the BioBrick prefix (brown) & suffix (purple) as well as a LIC overhang sequence (pink). | ||
| + | |||
{| | {| | ||
| - | |width="2000px" align="center"|[[Image:gen_gene_LICprimed.png|LIC-primed gene| | + | |width="2000px" align="center"|[[Image:gen_gene_LICprimed.png|LIC-primed gene|350px]] |
|- | |- | ||
| align="center" size="large" | '''↓''' | | align="center" size="large" | '''↓''' | ||
| Line 44: | Line 53: | ||
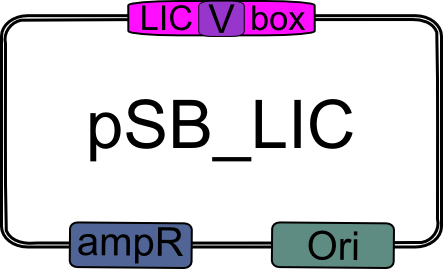
| - | 3.) Obtain LIC plasmid (pSB_LIC; part [http://partsregistry.org/Part:BBa_K155000 K155000]). | + | ==== 3.) LIC plasmid ==== |
| + | |||
| + | Obtain LIC plasmid (pSB_LIC; part [http://partsregistry.org/Part:BBa_K155000 K155000]). | ||
| + | |||
{| | {| | ||
| - | |width="2000px" align="center"|[[Image:pSB_LIC.png|pSB_LIC| | + | |width="2000px" align="center"|[[Image:pSB_LIC.png|pSB_LIC|200px]] |
|} | |} | ||
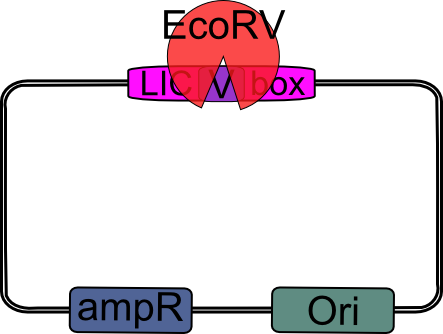
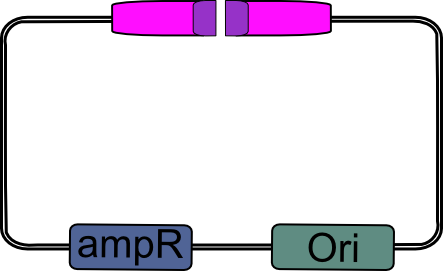
| - | 4.) Digest pSB_LIC with ''Eco''RV. | + | ==== 4.) EcoRV digestion ==== |
| + | |||
| + | Digest pSB_LIC with ''Eco''RV. | ||
| + | |||
{| | {| | ||
| - | |width="2000px" align="center"|[[Image:pSB_LIC_EcoRV.png|pSB_LIC digest with EcoRV| | + | |width="2000px" align="center"|[[Image:pSB_LIC_EcoRV.png|pSB_LIC digest with EcoRV|200px]] |
|- | |- | ||
| - | |width="2000px" align="center"|[[Image:pSB_LIC_EcoRV_cut.png|pSB_LIC digested with EcoRV| | + | | align="center" size="large" | '''↓''' |
| + | |- | ||
| + | |width="2000px" align="center"|[[Image:pSB_LIC_EcoRV_cut.png|pSB_LIC digested with EcoRV|200px]] | ||
| + | |- | ||
| + | | align="center" size="large" | '''↓''' | ||
|- | |- | ||
| - | |width="2000px" align="center"|[[Image:pSB_LIC_lin.png|pSB_LIC digested with EcoRV (linear)| | + | |width="2000px" align="center"|[[Image:pSB_LIC_lin.png|pSB_LIC digested with EcoRV (linear)|500px]] |
|} | |} | ||
| - | 5.) Digest pSB_LIC with ''' | + | |
| + | ==== 5.) T4 Polymerase digestion ==== | ||
| + | |||
| + | Digest linearized pSB_LIC, along with desired insert sequence, with T4 DNA polymerase. This digestion is done in the presence of only one dNTP, complementary to a base absent from the LIC overhang. Therefore, polymerase activity is impossible, and the 3' - 5' exonuclease activity of T4 DNA polymerase predominates. The 3' end of each DNA strand is chewed away, until an appropriate base is reached at the end of the LIC overhang. The dNTP required to base-pair with this base is the one added to the digestion buffer, and so the polymerase activity now balances the exonuclease activity, ending net digestion. | ||
| + | |||
{| | {| | ||
| - | |width="2000px" align="center"|[[Image: | + | |width="2000px" align="center"|[[Image:pSB_LIC_post_T4.png|linearized pSB_LIC digested with T4 polymerase|500px]] |
| + | |- | ||
| + | | align="center" size="large" | and | ||
| + | |- | ||
| + | |width="2000px" align="center"|[[Image:gen_gene_postT4.png|gene of interest digested with T4 polymerase|400px]] | ||
|} | |} | ||
| - | === ''' | + | ==== 6.) Mix Plasmid & Insert ==== |
| + | |||
| + | Mix T4 polymerase-treated pSB_LIC and insert, allowing complementary LIC overhangs to base-pair. The long base-paired regions are sufficient to hold them together through transformation, after which ''E. coli'' 's own ligase will repair the DNA backbone nicks. | ||
| + | |||
| + | {| | ||
| + | |width="2000px" align="center"|[[Image:gen_gene_in_pLIC.png|gene of interest in pSB_LIC|400px]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==== 7.) Transform & Use BioBrick==== | ||
| + | |||
| + | The DNA sequence of interest, with standard BioBrick prefix and suffix, is now ready for further use. | ||
| + | |||
| + | == '''Preparative Work''' == | ||
| + | |||
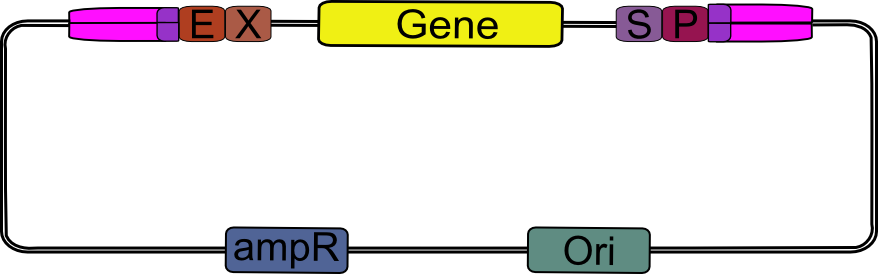
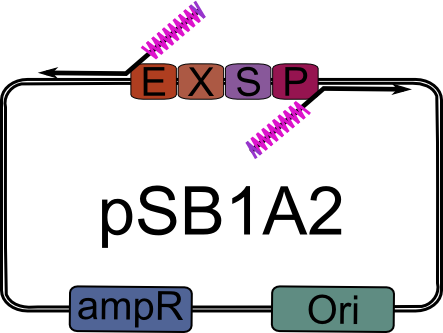
| + | We obtained pSB_LIC (part [http://partsregistry.org/Part:BBa_K155000 K155000]) by round-the-world PCR amplification from [http://partsregistry.org/Part:pSB1A2 pSB1A2], using a high-fidelity DNA polymerase. | ||
| + | |||
| + | {| | ||
| + | |width="2000px" align="center"|[[Image:pSB1A2_LIC_primed.png|pSB1A2 ready for pSB_LIC preparation|200px]] | ||
| + | |- | ||
| + | | align="center" size="large" | ↓ | ||
| + | |- | ||
| + | |width="2000px" align="center"|[[Image:pSB_LIC_lin.png|linear pSB_LIC|400px]] | ||
| + | |} | ||
| + | |||
| + | The linear PCR product was then blunt-end ligated to yield the circular plasmid submitted as part [http://partsregistry.org/Part:BBa_K155000 K155000]. | ||
| + | {| | ||
| + | |width="2000px" align="center"|[[Image:pSB_LIC.png|pSB_LIC|200px]] | ||
| + | |} | ||
| + | |||
| + | ==== Problem ==== | ||
| + | NB. Our intent was to use [http://partsregistry.org/Part:pSB1A7 pSB1A7], but the sample from the registry supposed to contain it, didn't. | ||
| + | |||
| + | == '''Intended Usage''' == | ||
| + | |||
| + | We used a complete GFP sequence for a proof of principle of our method. We also attempted to create an xylE BioBrick for the [https://2008.igem.org/Team:University_of_Lethbridge University of Lethbridge team]. | ||
| + | |||
| + | == '''Reference''' == | ||
Aslanidis, C. & de Jong P.J. (1990) “Ligation-independent cloning of PCR products (LIC-PCR).” ''Nucl. Acids Res.'' '''18''':20, 6069-6074. | Aslanidis, C. & de Jong P.J. (1990) “Ligation-independent cloning of PCR products (LIC-PCR).” ''Nucl. Acids Res.'' '''18''':20, 6069-6074. | ||
| + | <!-- | ||
Plasmid: [http://partsregistry.org/Part:BBa_K155000 K155000] | Plasmid: [http://partsregistry.org/Part:BBa_K155000 K155000] | ||
| + | --> | ||
<!-- | <!-- | ||
Latest revision as of 03:30, 30 October 2008
Our project centers on the development and optimization of a ligase-independent cloning (LIC) protocol.
Contents |
Abstract
Ligase-Independent Cloning as a Standard for BioBrick Preparation
While there is an established BioBrick format, there is not yet a standard method for turning a gene of interest into a BioBrick. Ideally, such a standard method would be easily adopted, even by amateurs, and would lend itself to automation. A significant drawback of several existing techniques is their dependence on ligase treatment, which is often problematic. We propose a ligase-independent cloning (LIC) method, based on the technique of [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)], as a possible standard for novel BioBrick preparation. Instead of short overhangs and ligase treatment, LIC uses long overhangs to circularize plasmid vectors for transformation without the use of ligase. The LIC method reduces the number of enzyme steps required for cloning, thus lending itself to easy adoption, automation, and real biological 'engineering.'
Rationale
The underlying idea of iGEM is that an engineering approach can be applied to biology. Using a computer analogy, the goal is to have software (DNA sequences) that can be run on compatible hardware (microorganisms) anywhere in the world.
| | 
| |
| + | = | + |
 | 
|
The portability of such a biological hardware-software interface requires extensive standardization. The requisite standards exist for the definition of BioBricks, and for the use of BioBrick parts in composite devices. There is not yet a standard method, though, for constructing new BioBricks.
Ideally, whatever standard method is chosen would not only be highly portable, but also simple. The BioBricks used by iGEM are advertised with Lego® blocks, implying a very easy-to-use set of tools. Our team of high school students discovered that synthetic biology is not (yet, at least) at the stage of quick and easy implementation in just anyone's kitchen or garage.
Therefore, we chose to investigate ligase-independent cloning (LIC), a technique we believe can serve as a standard for BioBrick construction, in a way that is relatively easy to implement and that also lends itself to automation in the future. Our approach was based on a method described by [http://nar.oxfordjournals.org/cgi/content/abstract/18/20/6069 Aslanidis & de Jong (1990)].
Method
The intent is to be able to create a standard BioBrick from any DNA sequence of interest.
1.) Select Gene
Identify a gene (DNA sequence) of interest
2.) Amplify Gene
Amplify, using PCR primers that contain the BioBrick prefix (brown) & suffix (purple) as well as a LIC overhang sequence (pink).

|
| ↓ |
3.) LIC plasmid
Obtain LIC plasmid (pSB_LIC; part [http://partsregistry.org/Part:BBa_K155000 K155000]).
|
4.) EcoRV digestion
Digest pSB_LIC with EcoRV.
|
| ↓ |
|
| ↓ |
5.) T4 Polymerase digestion
Digest linearized pSB_LIC, along with desired insert sequence, with T4 DNA polymerase. This digestion is done in the presence of only one dNTP, complementary to a base absent from the LIC overhang. Therefore, polymerase activity is impossible, and the 3' - 5' exonuclease activity of T4 DNA polymerase predominates. The 3' end of each DNA strand is chewed away, until an appropriate base is reached at the end of the LIC overhang. The dNTP required to base-pair with this base is the one added to the digestion buffer, and so the polymerase activity now balances the exonuclease activity, ending net digestion.
| and |
6.) Mix Plasmid & Insert
Mix T4 polymerase-treated pSB_LIC and insert, allowing complementary LIC overhangs to base-pair. The long base-paired regions are sufficient to hold them together through transformation, after which E. coli 's own ligase will repair the DNA backbone nicks.
|
7.) Transform & Use BioBrick
The DNA sequence of interest, with standard BioBrick prefix and suffix, is now ready for further use.
Preparative Work
We obtained pSB_LIC (part [http://partsregistry.org/Part:BBa_K155000 K155000]) by round-the-world PCR amplification from [http://partsregistry.org/Part:pSB1A2 pSB1A2], using a high-fidelity DNA polymerase.
|
| ↓ |
The linear PCR product was then blunt-end ligated to yield the circular plasmid submitted as part [http://partsregistry.org/Part:BBa_K155000 K155000].

|
Problem
NB. Our intent was to use [http://partsregistry.org/Part:pSB1A7 pSB1A7], but the sample from the registry supposed to contain it, didn't.
Intended Usage
We used a complete GFP sequence for a proof of principle of our method. We also attempted to create an xylE BioBrick for the University of Lethbridge team.
Reference
Aslanidis, C. & de Jong P.J. (1990) “Ligation-independent cloning of PCR products (LIC-PCR).” Nucl. Acids Res. 18:20, 6069-6074.
Main Page | Team | Project | Lab Notebook
 "
"