Team:Paris/constructions
From 2008.igem.org
(→Model constructions: from the modelling to the characterization) |
|||
| (9 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
Our project BacterioClock is based on an oscillating FIFO synchronized at the population level. To obtain and have preliminary results of this system we divided it in three "little" modules as the modeling team exposed them previously. | Our project BacterioClock is based on an oscillating FIFO synchronized at the population level. To obtain and have preliminary results of this system we divided it in three "little" modules as the modeling team exposed them previously. | ||
| - | + | ==The First one aims to see the FIFO which constitute our core system== | |
A simple manner to see it, is to implement one class II promotor followed by a reporter gene like GFP in a bacteria strain that is able to produce the flagella. | A simple manner to see it, is to implement one class II promotor followed by a reporter gene like GFP in a bacteria strain that is able to produce the flagella. | ||
| - | + | ||
By this way, we are able to see in single cell the order of the activation of pFliL then pFlgA then pFlhB and then the inactivation in the same order as a FIFO will do. | By this way, we are able to see in single cell the order of the activation of pFliL then pFlgA then pFlhB and then the inactivation in the same order as a FIFO will do. | ||
These constructions are equivalent to the experiments realized by Uri Alon in the article "Using quantitative Blueprint to reprogram the Dynamics of the Flagella Gene Network" Kalir S, Alon U. Cell 2004. | These constructions are equivalent to the experiments realized by Uri Alon in the article "Using quantitative Blueprint to reprogram the Dynamics of the Flagella Gene Network" Kalir S, Alon U. Cell 2004. | ||
| - | We are extremely grateful for Alon U. that send us the inducible gene ''FlhDC'', and ''FliA'' in pBad18 plasmid. | + | A better way to observe the FIFO is to study the system with an inducible regulator of class II gene in a bacteria strain deleted for this gene. We are extremely grateful for Alon U. that send us the inducible gene ''FlhDC'', and ''FliA'' in pBad18 plasmid. |
| - | These plasmids with our own araC/pBad-EnvZ* will allowed us to study the system | + | These plasmids with our own araC/pBad-EnvZ* will allowed us to study the system in mutated strains that we have in our lab (ΔFlhD, ΔFliA, ΔFlgM, ΔEnvZ). By cotransforming in mutated strains the inducibles regulators of the class II promotors with one of this promotor associated to a fluorescent protein, we could characterize the influence of the master regulators of the flagella on their pormoter. The fluorescence is normalized to the OD<sub>600</sub>. |
| - | For example: | + | <br>For example below are some constructions to see and induce the FIFO: |
| - | + | <br><br><br> | |
| - | + | [[Image:Felipe01.jpg|500px|center|thumb|pFliL alone]]<br><br><br> | |
| + | [[Image:Felipe02.jpg|500px|center|thumb|3 promoters of the FIFO in one Cell]]<br><br><br> | ||
| + | [[Image:Felipe03.jpg|500px|center|thumb|FIFO and oscillating construction in a single cell]]<br><br><br> | ||
| Line 22: | Line 24: | ||
One problem that could occur is the interaction between the differents fluorescents proteins due to the cross-over of their emission and excitation spectra and moreover in our final system that count three fluorophores. | One problem that could occur is the interaction between the differents fluorescents proteins due to the cross-over of their emission and excitation spectra and moreover in our final system that count three fluorophores. | ||
| - | + | [[Image:Spectre.jpg |600px |thumb| emission and excitation spectra of ECFP, YFP and mRFP]] | |
| - | The choice of ECFP, YFP and mRFP is based upon this consideration and upon the order of appearing in the FIFO. | + | <br>The choice of ECFP, YFP and mRFP is based upon this consideration and upon the order of appearing in the FIFO. |
Indeed by putting the mRFP in last position, we improve the visualization of the FIFO like the maturation time of the protein is about of one hour. | Indeed by putting the mRFP in last position, we improve the visualization of the FIFO like the maturation time of the protein is about of one hour. | ||
| - | We thank a lot Arnau Montagu advisor | + | We thank a lot Arnau Montagu, advisor of the iGEM Valencia Team. He advised us, after his team had troubleshooting with fluorescence, to take care about ''spurious FRET''. This phenomenon occurs even with non covered fluorescent spectra when the concentration of two fluorophores is sufficient so they are enough closer to induce some FRET. But it could happen at lower concentration than 40 μM. <br> |
<br>- "DsRed as a Potential FRET Partner with CFP and GFP" Michael G. Erickson, Daniel L. Moon, and David T. Yue Biophysical Journal 2003. | <br>- "DsRed as a Potential FRET Partner with CFP and GFP" Michael G. Erickson, Daniel L. Moon, and David T. Yue Biophysical Journal 2003. | ||
<br>- "A Comparison of Donor-Acceptor Pairs for Genetically Encoded FRET Sensors: Application to the Epac cAMP | <br>- "A Comparison of Donor-Acceptor Pairs for Genetically Encoded FRET Sensors: Application to the Epac cAMP | ||
Sensor as an Example" Gerard N. M. van der Krogt, Janneke Ogink, Bas Ponsioen, Kees Jalink PLoS 2008. | Sensor as an Example" Gerard N. M. van der Krogt, Janneke Ogink, Bas Ponsioen, Kees Jalink PLoS 2008. | ||
| + | |||
| + | <br> | ||
| + | ==Oscillating module== | ||
| + | Few approaches have been thought in order to create the oscillating module. The pTet/TetR system that allows us to precisely control the inhibition of TetR on the Tet promotor. Another system use the natural inhibition of OmpR and of EnvZ via OmpR. We finally perform this system that we have developed since we know thanks to the modeling team that the oscillations are the only way to see the full development of the FIFO. | ||
Latest revision as of 03:42, 30 October 2008
Model constructions: from the modelling to the characterization
Our project BacterioClock is based on an oscillating FIFO synchronized at the population level. To obtain and have preliminary results of this system we divided it in three "little" modules as the modeling team exposed them previously.
The First one aims to see the FIFO which constitute our core system
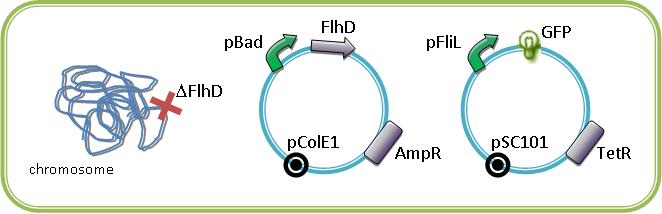
A simple manner to see it, is to implement one class II promotor followed by a reporter gene like GFP in a bacteria strain that is able to produce the flagella.
By this way, we are able to see in single cell the order of the activation of pFliL then pFlgA then pFlhB and then the inactivation in the same order as a FIFO will do.
These constructions are equivalent to the experiments realized by Uri Alon in the article "Using quantitative Blueprint to reprogram the Dynamics of the Flagella Gene Network" Kalir S, Alon U. Cell 2004.
A better way to observe the FIFO is to study the system with an inducible regulator of class II gene in a bacteria strain deleted for this gene. We are extremely grateful for Alon U. that send us the inducible gene FlhDC, and FliA in pBad18 plasmid.
These plasmids with our own araC/pBad-EnvZ* will allowed us to study the system in mutated strains that we have in our lab (ΔFlhD, ΔFliA, ΔFlgM, ΔEnvZ). By cotransforming in mutated strains the inducibles regulators of the class II promotors with one of this promotor associated to a fluorescent protein, we could characterize the influence of the master regulators of the flagella on their pormoter. The fluorescence is normalized to the OD600.
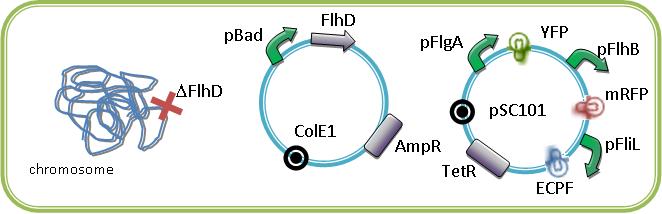
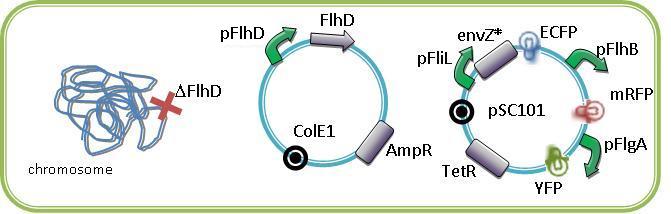
For example below are some constructions to see and induce the FIFO:
To perform such a cotransformation we take care about the ORI of each low copy plasmid which are often incompatible and we made all our final constructions in the pSB4T5 plasmid that care the only one resistance that was not already used and the pSC101 ORI which is compatible with the ColE1.
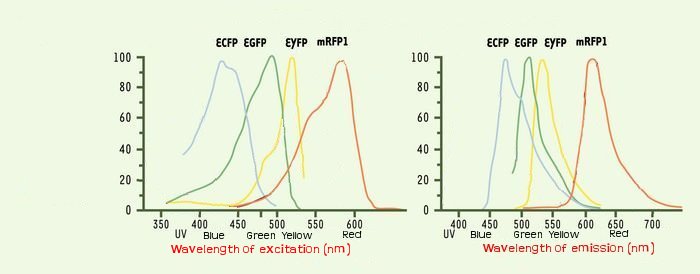
We could see the FIFO in a more convenient manner with the assembly of two class II promotors associated to differents fluorophores. Then we could observed two kinds of fluorescents in One Cell along time. One problem that could occur is the interaction between the differents fluorescents proteins due to the cross-over of their emission and excitation spectra and moreover in our final system that count three fluorophores.
The choice of ECFP, YFP and mRFP is based upon this consideration and upon the order of appearing in the FIFO.
Indeed by putting the mRFP in last position, we improve the visualization of the FIFO like the maturation time of the protein is about of one hour.
We thank a lot Arnau Montagu, advisor of the iGEM Valencia Team. He advised us, after his team had troubleshooting with fluorescence, to take care about spurious FRET. This phenomenon occurs even with non covered fluorescent spectra when the concentration of two fluorophores is sufficient so they are enough closer to induce some FRET. But it could happen at lower concentration than 40 μM.
- "DsRed as a Potential FRET Partner with CFP and GFP" Michael G. Erickson, Daniel L. Moon, and David T. Yue Biophysical Journal 2003.
- "A Comparison of Donor-Acceptor Pairs for Genetically Encoded FRET Sensors: Application to the Epac cAMP
Sensor as an Example" Gerard N. M. van der Krogt, Janneke Ogink, Bas Ponsioen, Kees Jalink PLoS 2008.
Oscillating module
Few approaches have been thought in order to create the oscillating module. The pTet/TetR system that allows us to precisely control the inhibition of TetR on the Tet promotor. Another system use the natural inhibition of OmpR and of EnvZ via OmpR. We finally perform this system that we have developed since we know thanks to the modeling team that the oscillations are the only way to see the full development of the FIFO.
 "
"