Team:Imperial College/Genetic Circuit
From 2008.igem.org
m |
m (Small grammar edits) |
||
| (One intermediate revision not shown) | |||
| Line 3: | Line 3: | ||
=== Modelling the Genetic Circuit === | === Modelling the Genetic Circuit === | ||
{{Imperial/Box2|| | {{Imperial/Box2|| | ||
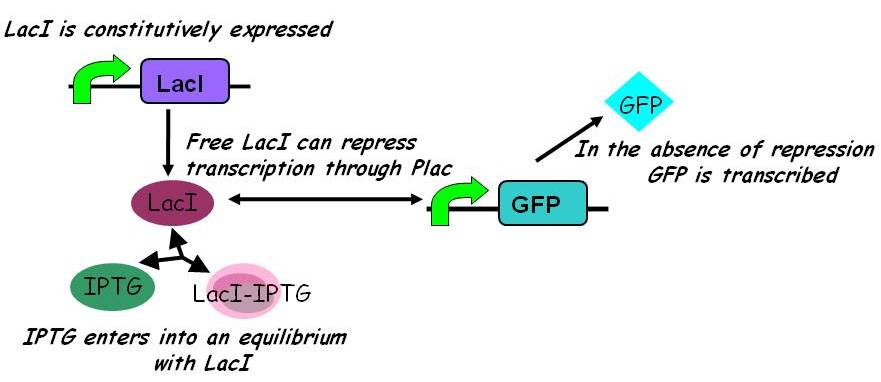
| - | An accurate mathematical description of the behaviour of | + | An accurate mathematical description of the behaviour of genetic circuits is essential for projects involving synthetic biology. Such descriptions are an integral component of part submission to the Registry, as exemplified by the canonical characterisation of part F2620 ({{ref|1}}). The ability to capture part behaviour as a mathematical relationship between input and output is useful for future re-use of the part and modification or integration into novel genetic circuits.}} |
{{Imperial/Box1|Modelling Constitutive Gene Expression| | {{Imperial/Box1|Modelling Constitutive Gene Expression| | ||
| Line 10: | Line 10: | ||
[[Image:Eq1.png]] | [[Image:Eq1.png]] | ||
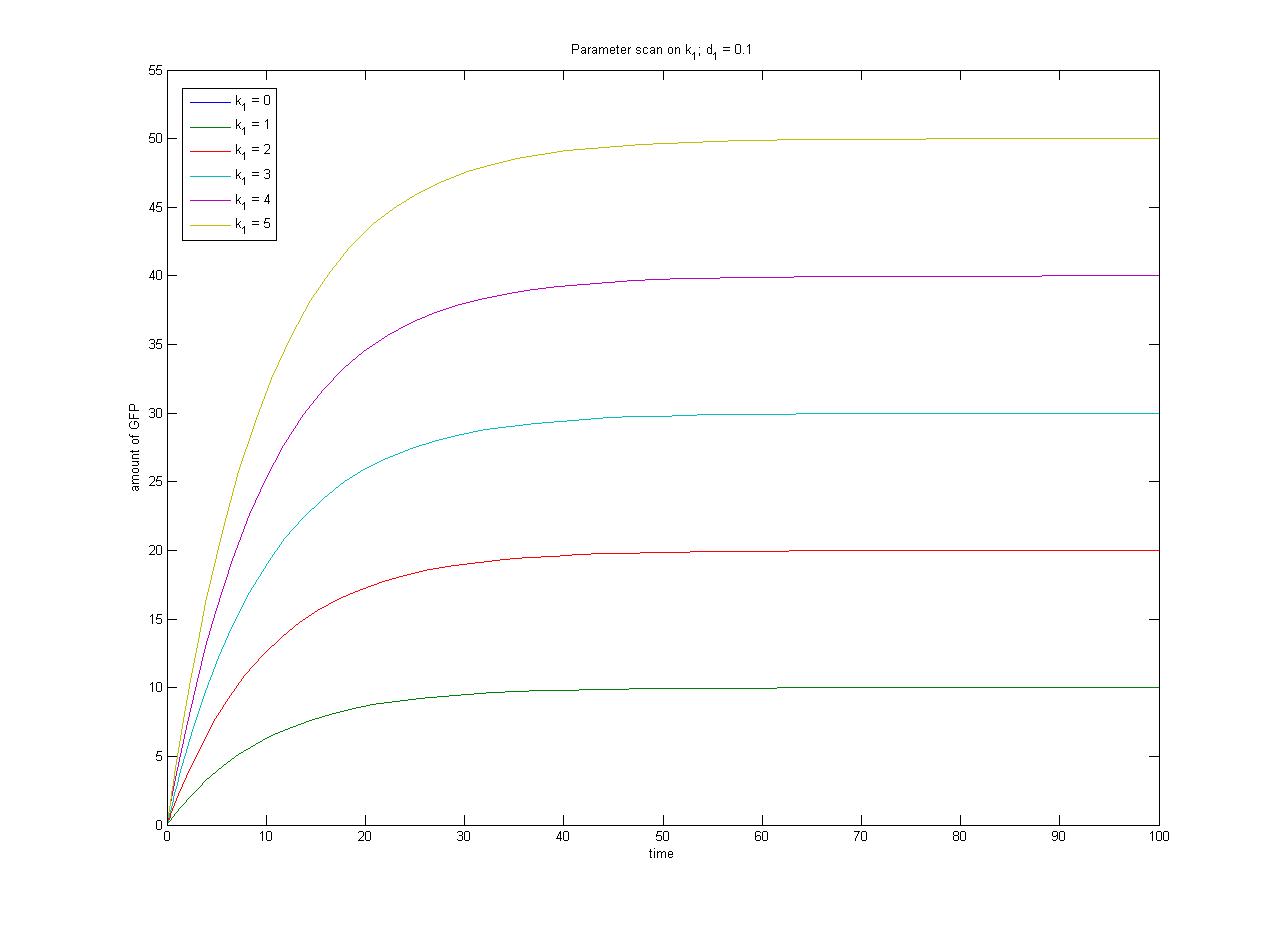
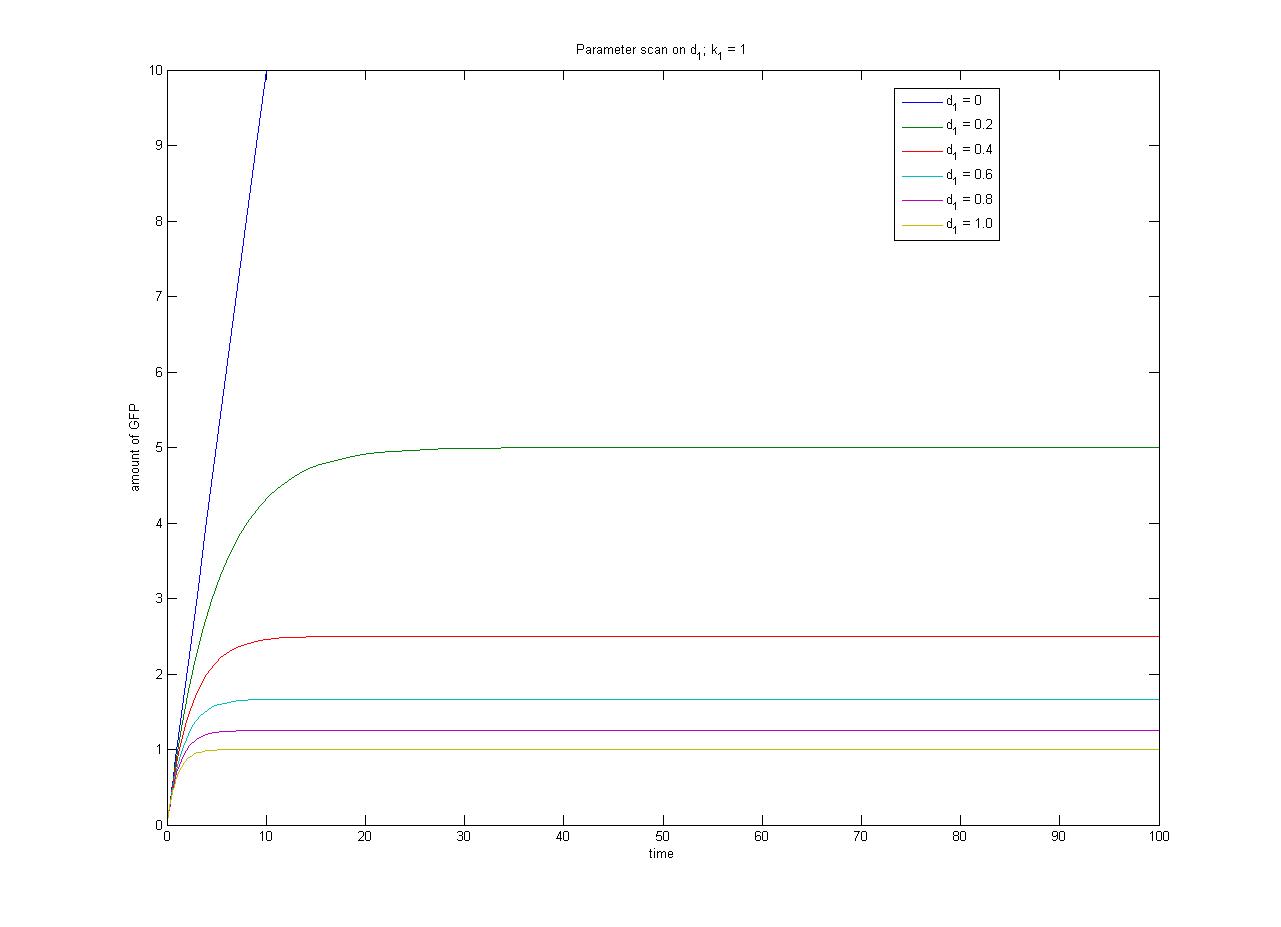
| - | In this case, [protein] represents the concentration of GFP, which will be the reporter protein in our constructs used for | + | In this case, [protein] represents the concentration of GFP, which will be the reporter protein in our constructs used for characterization. k<sub>1</sub> represents the rate of synthesis and d<sub>1</sub> represents the degradation rate. |
| - | We can easily simulate this synthesis-degradation model using Matlab; the ODEs and simulation file can be found in the | + | We can easily simulate this synthesis-degradation model using Matlab; the ODEs and simulation file can be found in the dry lab appendices. |
We can also solve this ODE analytically. | We can also solve this ODE analytically. | ||
| Line 59: | Line 59: | ||
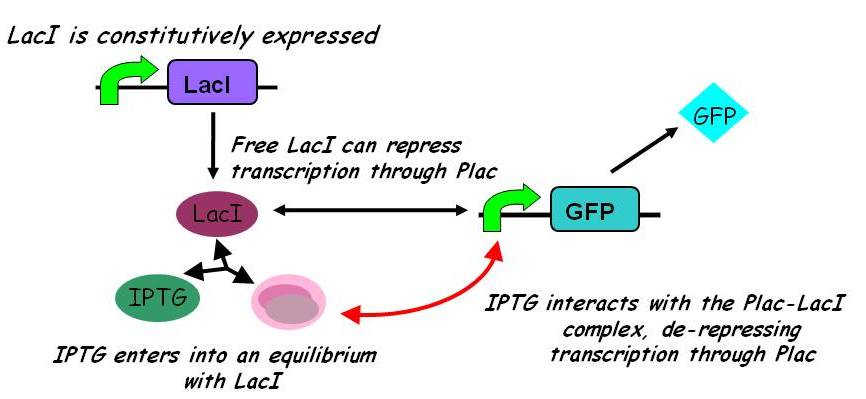
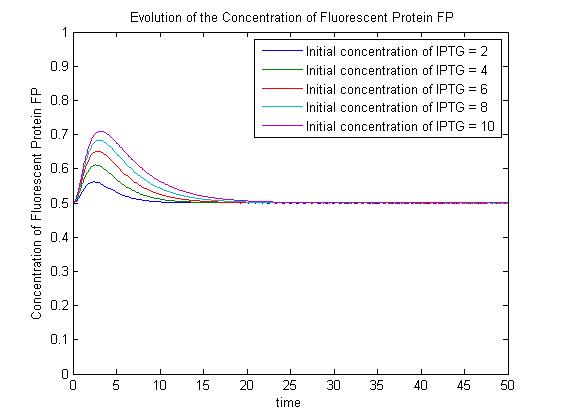
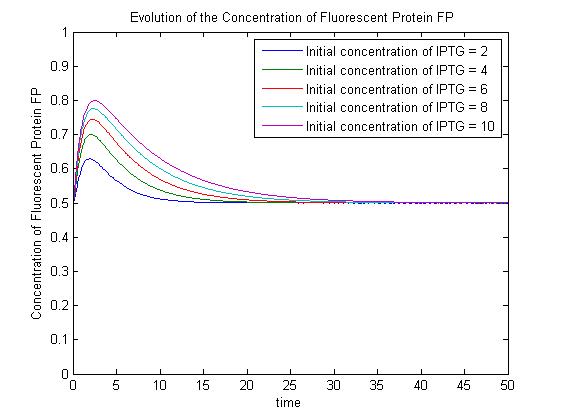
Under this model, the dynamic behaviour (whether or not [GFP] attains a maximum higher than its steady-state value) depends on the relative strengths of the kinetic constants describing the interactions underlying the model. Either way, the steady-state [GFP] will vary as a Hill-function dependent on the initial concentration of IPTG; this characteristic can be used to discriminate between the two models. | Under this model, the dynamic behaviour (whether or not [GFP] attains a maximum higher than its steady-state value) depends on the relative strengths of the kinetic constants describing the interactions underlying the model. Either way, the steady-state [GFP] will vary as a Hill-function dependent on the initial concentration of IPTG; this characteristic can be used to discriminate between the two models. | ||
| - | ODEs and simulation m-files for further exploration of the properties of these models can be found in the | + | ODEs and simulation m-files for further exploration of the properties of these models can, again, be found in the appendices. |
[[Team:Imperial_College/Genetic_Circuit_Details | '''>>> Details of equilibria and equations and qualitative discussion of parameter effects >>>''']] | [[Team:Imperial_College/Genetic_Circuit_Details | '''>>> Details of equilibria and equations and qualitative discussion of parameter effects >>>''']] | ||
Latest revision as of 04:03, 27 July 2009
Modelling the Genetic Circuit
|
|||||||||||||||||||
 "
"