Team:Chiba/Experiments:Reporter
From 2008.igem.org
(→Reporter) |
|||
| (22 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html><link rel="stylesheet" href="https://2008.igem.org/wiki/index.php?title=User:Maiko/chiba.css&action=raw&ctype=text/css" type="text/css" /></html> | <html><link rel="stylesheet" href="https://2008.igem.org/wiki/index.php?title=User:Maiko/chiba.css&action=raw&ctype=text/css" type="text/css" /></html> | ||
[[Image:Chiba-U.gif|center]] | [[Image:Chiba-U.gif|center]] | ||
| - | {| style="color:white; | + | {| style="color:white;" cellpadding="3" cellspacing="3" border="0" width="100%" align="center" class="menu" | |
!align="center"|[[Team:Chiba|Home]] | !align="center"|[[Team:Chiba|Home]] | ||
!align="center"|[[Team:Chiba/Team|The Team]] | !align="center"|[[Team:Chiba/Team|The Team]] | ||
| Line 13: | Line 13: | ||
==Reporter== | ==Reporter== | ||
| + | Our delayed switch has to turn on or off without variable time lag | ||
| + | after the preset time has passed. Therefore we need the output module, | ||
| + | which announces quickly that a particular concentration of the | ||
| + | signaling molcules has been reached. | ||
| + | We imagined three kinds of output: | ||
| - | |||
| - | + | * Fluorescent Protein | |
| + | * Bioluminescence(luciferase) | ||
| + | * Cell staining (LacZ and X-gal assay) | ||
| - | |||
| - | |||
| - | |||
| + | and tested Fluorescent Protein and β-gal(lacZ). | ||
| - | We | + | We selected GFP, Venus YFP, and mCherry. The latter two were chosen because Venus YFP matures quickly[[Team:Chiba/Reference#Output|(1)]],and we can easibly recognize mCherry coloration[[Team:Chiba/Reference#Output|(2)]]. |
| + | We used fluorescent proteins and β-gal(lacZ) as a reporter. | ||
| - | |||
| - | + | We were able to recognize chemical luminescence of luciferase only at | |
| - | + | dark place devoid of light. Therefore the conditions of use are | |
| - | + | limited, and we did not select Luciferase as our reporter. | |
==Experiment== | ==Experiment== | ||
| - | |||
| - | |||
| + | Measure the time course form adding inducer (IPTG or AHL) to confirm the change of color to analysis the gene expression. | ||
| + | Evaluate following gene: | ||
| - | + | *Fluorescent Protein | |
| - | + | :*GFP | |
| - | + | ::*pGFPuv | |
| + | ::[[Image:PGFPuv chiba.gif]] | ||
| + | ::*[http://partsregistry.org/Part:BBa_T9002 BBa_T9002] | ||
| + | ::[[Image:High-Copy-Receiver Chiba.gif]] | ||
| + | :*Venus YFP | ||
| + | ::*[http://partsregistry.org/wiki/index.php?title=Part:BBa_K084003 BBa_K084003] | ||
| + | ::[[Image:K084003 chiba.gif]] | ||
| + | ::*pLac-Venus YFP | ||
| + | ::[[Image:pLac_Venus_Chiba.gif]] | ||
| + | :*mCherry | ||
| + | ::*pLac-mCherry | ||
| + | ::[[Image:PLac_mChe_Chiba.gif]] | ||
| + | *β-gal (X-gal assay) | ||
| + | :*pUC19(plac-LacZα) | ||
| + | ::[[Image:PLac-lacZ-Chiba.gif]] | ||
*Fluorescent Protein | *Fluorescent Protein | ||
| Line 66: | Line 84: | ||
'''Result''' | '''Result''' | ||
'''LacZ''' | '''LacZ''' | ||
| - | [[Image:LacZ chiba.gif|thumb|right|'''Table | + | [[Image:LacZ chiba.gif|thumb|right|'''Table 1''' Dependent on X-gal concentration.]] |
| - | * | + | * LacZ, which is not fluorescent, was observed under the naked eye. |
| - | * | + | * Time required for observation of LacZ staining depended on X-gal concentration. |
| - | * | + | * We were not able to observe stainig at a liquid state within 4 hours.-->Staining was observed at the 8 hour point. |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | '''Fluorescent protein''' | ||
| + | [[Image:Reporter chiba.gif|thumb|right|'''Table 2''' Result of Time chase.]] | ||
| + | * Similar results were obtained for liquid and solid cultures. | ||
| + | * To the naked eye, fluorescence was observed most easily in the order mCherry, GFP, and YFP. | ||
| + | * The solid state cultures were easier to observe to the naked eye. | ||
| + | * AHL induction took a shorter time than IPTG for fluorescence to be observable. | ||
| + | * GFP and YFP showed observable fluorescence earlier than mCherry. | ||
<br clear=all> | <br clear=all> | ||
| - | [[Image:Soliduv Chiba.gif|thumb|right|''' | + | [[Image:Soliduv Chiba.gif|thumb|right|'''Photo 1''' Emission of fluorescent protein]] |
'''Discussion''' | '''Discussion''' | ||
| + | Discussion | ||
| - | |||
| - | + | The purpose of this project is to alter the time required for expression. | |
| - | *X- | + | * By increasing the concentration of X-gal, the time required for |
| + | expression to be observable decreased, but this still takes longer | ||
| + | than the time required when using fluorescent proteins. | ||
| - | |||
| - | + | *Using β-gal, which uses X-gal as substrate, the time required for | |
| + | output may increase because the substrate concentration decreases with | ||
| + | time. | ||
| - | * | + | For the above reasons, fluorescence proteins are more suited for our purposes. |
| + | |||
| + | Of the fluorescent proteins, GFP and YFP fluorescence was observable | ||
| + | at earlier stages. | ||
| + | |||
| + | |||
| + | * But to the naked eye, GFP could be observed more easily. | ||
| + | |||
| + | Therefore, we chose GFP as our output reporter. | ||
| - | |||
<br clear=all> | <br clear=all> | ||
===Method=== | ===Method=== | ||
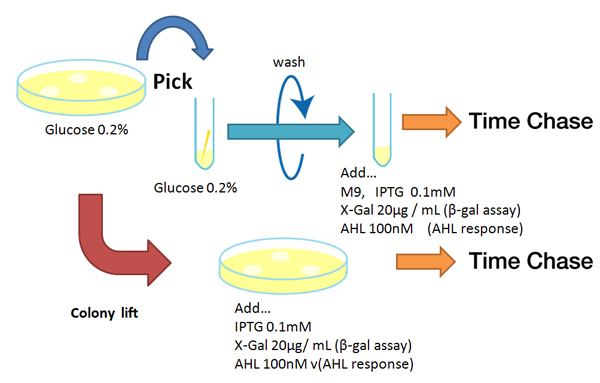
| - | [[Image:Reporter method Chiba.jpg|rifht|thumb|'''Fig. ''' | + | [[Image:Reporter method Chiba.jpg|rifht|thumb|'''Fig. 1''' Experimental method for response measuring of reporters]] |
Strain:XL10G Kan<sup>R</sup> | Strain:XL10G Kan<sup>R</sup> | ||
*Agar plate experiment | *Agar plate experiment | ||
| Line 153: | Line 180: | ||
:Beckman Allegratm X-12R Centrifuga(Beckman Coulter) | :Beckman Allegratm X-12R Centrifuga(Beckman Coulter) | ||
| - | + | {| style="color:white;" cellpadding="3" cellspacing="3" border="0" width="100%" align="center" class="menu" | | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| style="color:white; | + | |
!align="center"|[[Team:Chiba|Home]] | !align="center"|[[Team:Chiba|Home]] | ||
!align="center"|[[Team:Chiba/Team|The Team]] | !align="center"|[[Team:Chiba/Team|The Team]] | ||
Latest revision as of 12:43, 16 December 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Reporter
Our delayed switch has to turn on or off without variable time lag after the preset time has passed. Therefore we need the output module, which announces quickly that a particular concentration of the signaling molcules has been reached.
We imagined three kinds of output:
- Fluorescent Protein
- Bioluminescence(luciferase)
- Cell staining (LacZ and X-gal assay)
and tested Fluorescent Protein and β-gal(lacZ).
We selected GFP, Venus YFP, and mCherry. The latter two were chosen because Venus YFP matures quickly(1),and we can easibly recognize mCherry coloration(2).
We used fluorescent proteins and β-gal(lacZ) as a reporter.
We were able to recognize chemical luminescence of luciferase only at
dark place devoid of light. Therefore the conditions of use are
limited, and we did not select Luciferase as our reporter.
Experiment
Measure the time course form adding inducer (IPTG or AHL) to confirm the change of color to analysis the gene expression.
Evaluate following gene:
- Fluorescent Protein
- GFP
- Venus YFP
- mCherry
- β-gal (X-gal assay)
- Fluorescent Protein
- GFP
- Venus YFP
- mCherry
- β-gal (X-gal assay)
Result & Discussion
Result LacZ
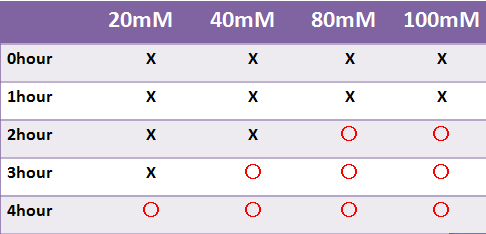
- LacZ, which is not fluorescent, was observed under the naked eye.
- Time required for observation of LacZ staining depended on X-gal concentration.
- We were not able to observe stainig at a liquid state within 4 hours.-->Staining was observed at the 8 hour point.
Fluorescent protein
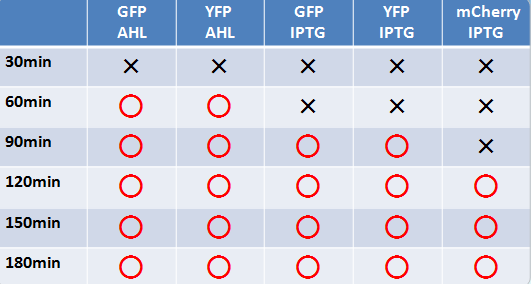
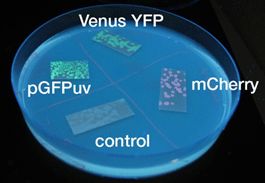
- Similar results were obtained for liquid and solid cultures.
- To the naked eye, fluorescence was observed most easily in the order mCherry, GFP, and YFP.
- The solid state cultures were easier to observe to the naked eye.
- AHL induction took a shorter time than IPTG for fluorescence to be observable.
- GFP and YFP showed observable fluorescence earlier than mCherry.
Discussion Discussion
The purpose of this project is to alter the time required for expression.
- By increasing the concentration of X-gal, the time required for
expression to be observable decreased, but this still takes longer than the time required when using fluorescent proteins.
- Using β-gal, which uses X-gal as substrate, the time required for
output may increase because the substrate concentration decreases with time.
For the above reasons, fluorescence proteins are more suited for our purposes.
Of the fluorescent proteins, GFP and YFP fluorescence was observable at earlier stages.
- But to the naked eye, GFP could be observed more easily.
Therefore, we chose GFP as our output reporter.
Method
Strain:XL10G KanR
- Agar plate experiment
- Pre-culture
- Picked and cultured the following plate stocks in 2mL of LB:
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Picked and cultured the following plate stocks in 2mL of LB:
- Spread on plate
- Spread on new plate
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Spread on new plate
- Colony lift
- Colony lift to inducible agar plate (containing IPTG or AHL)
- LB-Amp+0.2 mM IPTG agar plate, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp+100 nM AHL agar plate, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Incubate at 37 °C
- Colony lift to inducible agar plate (containing IPTG or AHL)
- Check expression every 30 min.
- Liquid medium experiment
- Pre-culture
- Picked and cultured the following plate in 2mL of LB:
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for 12h.
- Picked and cultured the following plate in 2mL of LB:
- Culture
- Dilute pre-cultures and add to new LB medium.
- LB-Amp+0.2 % Glucose, (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- LB-Amp, ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Cultured at 37°C for about 6 h
- Dilute pre-cultures and add to new LB medium.
- Wash
- Transfer 10mL each of the culture to 50mL centrifuge tubes.
- Centrifuged for 6min at 3600rpm,20°C and discarded the supernatant.
- Add physiological saline and resuspention
- Repeated wash twice.
- Add M9 minimal medium.
- Mix
- Dispens each culture into 48-well deep well or 96-well deep well
- Add IPTG fainal conc. 0.2 mM (pGFPuv, pLac-Venus YFP, pLac-mCherry, pUC19)
- Add AHL fainal conc. 100nM ([http://partsregistry.org/Part:BBa_T9002 BBa_T9002], [http://partsregistry.org/Part:BBa_K084003 BBa_K084003])
- Measure fluorescence intensity every 1 h.
- Equipment
- shaking incubator
- Innova 4200 Benchtop or Floor-Stackable Incubator Shaker(37°C)
- 46-well plate(deep well)
- 96-well plate(deep well)
- Fluoroskan Ascent 2.5(program:Ascent Software Version 2.6)
- Beckman Allegratm X-12R Centrifuga(Beckman Coulter)
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"