Team:University of Lethbridge/Modeling
From 2008.igem.org
David.franz (Talk | contribs) (Fixed section titles for inverted colors) |
(→Our Project) |
||
| (15 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{| align="center" style="background-color:#000000; color:white" | {| align="center" style="background-color:#000000; color:white" | ||
| - | !align="center"|[[Image:UofLteamlogo.jpg| | + | !align="center"|[[Image:UofLteamlogo.jpg|150px]] |
!align="center"|<span class="plainlinks" height:3px > | !align="center"|<span class="plainlinks" height:3px > | ||
| Line 29: | Line 29: | ||
</span> | </span> | ||
| - | !align="center"|[[Image:UofLteamlogo.jpg| | + | !align="center"|[[Image:UofLteamlogo.jpg|150px]] |
|} | |} | ||
==<span style="background-color:#000000; color:white">Our Project</span>== | ==<span style="background-color:#000000; color:white">Our Project</span>== | ||
| - | + | For our project, we decided to create a synthetic bacterium which could move towards and degrade an environmental pollutant. To direct the movement of our modified "Bacuum Cleaner," we reprogrammed the chemotaxis pathway by using a toxin-responsive riboswitch to control expression of the motility protein CheZ. CheZ catalyzes the dephosphorylation of the motility protein CheY, which promotes directed movement (chemotaxis) over the "tumbling" state of the bacterium. | |
| + | |||
| + | |||
| + | We used the modeling program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html <font color=yellow>BCT</font>] (Bray et al, 1993, 2007; Bray and Bourret, 1995) to verify that CheZ expression levels can be manipulated to control bacterial movement. The program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html <font color=yellow>BCT</font>] simulates the chemotaxis signalling pathway in ''Escherichia coli'' and is extensively supported by [http://www.pdn.cam.ac.uk/groups/comp-cell/Mutants.html <font color=yellow>experimental data</font>]. We used the program to demonstrate that controlling CheZ expression will control bacterial motility and to determine the amount of CheZ necessary to dephosphorylate CheY in high enough levels to produce directed movement. We modeled the E. coli strain RP437 in BCT, which is the same strain our group used as the chassis for our iGEM project. | ||
| + | |||
| + | |||
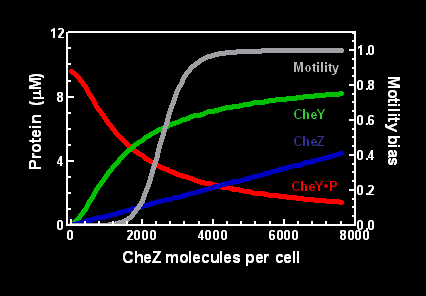
| + | Increasing intracellular amounts of CheZ led to increased concentration of CheY in the unphosphorylated state (Figure 1). Motility also increased, from a relative state of 0 (no movement or the tumbling state) to the relative state of 1 (continually moving in a directed manner). | ||
---- | ---- | ||
| - | |||
| + | [[Image:Chez_hj_black.gif|400px]] | ||
| + | <font color=white> Figure 1 | ||
| + | |||
| + | ==<span style="background-color:#000000; color:white">References</span>== | ||
| + | Bray, D., Bourret, R. B., and Simon, M. I. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. ''Mol. Biol. Cell'' 4, 469-482. | ||
| + | |||
| + | Bray, D., and Bourret, R. B. 1995. Computer analysis of the binding reactions leading to a transmembrane receptor-linked multiprotein complex involved in bacterial chemotaxis. ''Mol. Biol. Cell'' 6, 1367-1380. | ||
| + | |||
| + | Bray, D., Levin, M. D., and Lipkow, K. 2007. The chemotactic behavior of computer-based surrogate bacteria. ''Curr. Biol.'' 17, 12-19. | ||
| + | |||
| + | ---- | ||
{| align="center" style="background-color:#000000; color:white" | {| align="center" style="background-color:#000000; color:white" | ||
| - | !align="center"|[[Image:UofLteamlogo.jpg| | + | !align="center"|[[Image:UofLteamlogo.jpg|150px]] |
!align="center"|<span class="plainlinks" height:3px > | !align="center"|<span class="plainlinks" height:3px > | ||
| Line 69: | Line 85: | ||
</span> | </span> | ||
| - | !align="center"|[[Image:UofLteamlogo.jpg| | + | !align="center"|[[Image:UofLteamlogo.jpg|150px]] |
|} | |} | ||
Latest revision as of 01:00, 30 October 2008
Our Project
For our project, we decided to create a synthetic bacterium which could move towards and degrade an environmental pollutant. To direct the movement of our modified "Bacuum Cleaner," we reprogrammed the chemotaxis pathway by using a toxin-responsive riboswitch to control expression of the motility protein CheZ. CheZ catalyzes the dephosphorylation of the motility protein CheY, which promotes directed movement (chemotaxis) over the "tumbling" state of the bacterium.
We used the modeling program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html BCT] (Bray et al, 1993, 2007; Bray and Bourret, 1995) to verify that CheZ expression levels can be manipulated to control bacterial movement. The program [http://www.pdn.cam.ac.uk/groups/comp-cell/BCT.html BCT] simulates the chemotaxis signalling pathway in Escherichia coli and is extensively supported by [http://www.pdn.cam.ac.uk/groups/comp-cell/Mutants.html experimental data]. We used the program to demonstrate that controlling CheZ expression will control bacterial motility and to determine the amount of CheZ necessary to dephosphorylate CheY in high enough levels to produce directed movement. We modeled the E. coli strain RP437 in BCT, which is the same strain our group used as the chassis for our iGEM project.
Increasing intracellular amounts of CheZ led to increased concentration of CheY in the unphosphorylated state (Figure 1). Motility also increased, from a relative state of 0 (no movement or the tumbling state) to the relative state of 1 (continually moving in a directed manner).
Figure 1
References
Bray, D., Bourret, R. B., and Simon, M. I. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol. Biol. Cell 4, 469-482.
Bray, D., and Bourret, R. B. 1995. Computer analysis of the binding reactions leading to a transmembrane receptor-linked multiprotein complex involved in bacterial chemotaxis. Mol. Biol. Cell 6, 1367-1380.
Bray, D., Levin, M. D., and Lipkow, K. 2007. The chemotactic behavior of computer-based surrogate bacteria. Curr. Biol. 17, 12-19.
</div>
 "
"

 ]
]
 ]
]
 ]
]
 ]
]
 ]
]
 ]
]