Team:Illinois/Antibody RTK Fusion
From 2008.igem.org
(→Literature References) |
|||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:pcrpic.png|950px|center]] | |
| - | + | {{bottom_template}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Core Team Members== | ==Core Team Members== | ||
| Line 17: | Line 8: | ||
==Project Abstract== | ==Project Abstract== | ||
| - | In the diverse world of cellular | + | In the diverse world of cellular signaling pathways one of the most studied and well characterized family of receptors are the receptor tyrosine kinases. These cell surface proteins detect a wide range of extracellular ligands including hormones, growth factors, and cytokines. Receptor tyrosine kinases are most typically composed of an extracellular ligand binding domain, a single transmembrane helix, and an intracellular effector domain. Upon receptor binding of its ligand, signal cascades are activated within the cell. The first step in these signal cascades is often receptor oligomerization and cross-phosphorylation which leads to activation of MAP kinase cascades or other pathways. We plan to leverage the well understood mechanisms of receptor tyrosine kinases for the purpose of detecting the cholera toxin. |
==Specific Plans, Supplies, and Protocols== | ==Specific Plans, Supplies, and Protocols== | ||
| - | We have chosen yeast as our platform for this project due to it's ability to correctly express cell surface proteins. Yeast do not naturally produce receptor tyrosine kinases | + | We have chosen yeast as our platform for this project due to it's ability to correctly express cell surface proteins. Yeast do not naturally produce receptor tyrosine kinases, but mammalian RTKs have been functionally expressed in yeast. Our first step will be to express in yeast a well understood receptor tyrosine kinase, vascular endothelial growth factor receptor in this case. From here we will create a fusion protein that will allow us to detect a ligand of our choosing. We have chosen the cholera toxin, for which an antibody against the B subunit has been sequenced. A fusion protein containing this antibody in place of VEGFR's normal ligand binding domain will bind the cholera toxin, and due to the pentameric structure of the B subunit multiple receptors will be able to dimerize and thus activate a signal cascade. Natural antibodies are composed of four separate polypeptide chains, but it is not possible to fuse all of these chains to a receptor. To get around this problem, we will create a single-chain antibody which consists of the antigen binding fragments of the light and heavy chains of an antibody joined together by a flexible glycine-serine linker. This type of antibody has been shown to retain specificity and affinity comparable to native antibodies. Finally, we will integrate the signalling event created by antigen binding and receptor dimerization into an endogenous signaling pathway, which we will then engineer to activate transcription of a reporter such as GFP. |
| - | To | + | To implement this project we will need to acquire or clone the genes for our receptor tyrosine kinase, the cholera antibody, and other signaling proteins needed to interface with the yeast pathway. Techniques such as PCR, cloning, making fusion proteins, and yeast culture will be needed. Supplies needed will include the cholera B subunit, yeast culture media, and standard molecular biology reagents/kits. |
| - | This project can be easily modularized into several subprojects. Expression of the RTK can be carried out independently of ligand binding or signalling, and this could possibly be tested by using fluorescently tagged antibodies to check for cell surface localization of the RTK. The fusion protein could be constructed and antigen binding tested perhaps by FRET or some method of affinity chromatography. The signal cascade could be tested using the unmodified RTK and detecting whether our reporter gene is activated in the | + | This project can be easily modularized into several subprojects. Expression of the RTK can be carried out independently of ligand binding or signalling, and this could possibly be tested by using fluorescently tagged antibodies to check for cell surface localization of the RTK. The fusion protein could be constructed and antigen binding tested perhaps by FRET or some method of affinity chromatography. The signal cascade could be tested using the unmodified RTK and detecting whether our reporter gene is activated in the presence of the RTK's natural ligand. |
| - | == | + | ==In Pictures== |
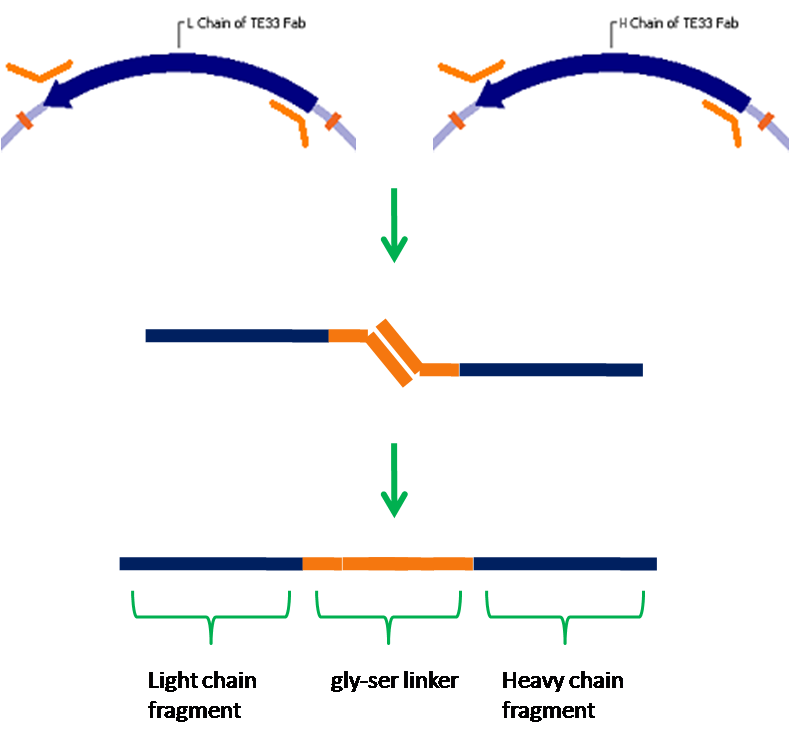
| - | + | [[Image:Assembly.png|left|400px|thumb|We have assembled a single chain antibody from the light and heavy antigen binding fragments of the TE33 antibody]] | |
| + | [[Image:Antibody.png|none|thumb|Generic antibody structure]] | ||
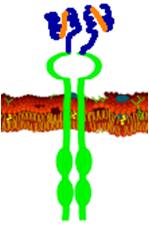
| + | [[Image:Artk.png|thumb|none|A schematic view of our antibody-receptor fusion. Two identical receptor monomers would be crosslinked by the cholera toxin and result in activation of the signal cascade.]] | ||
| - | |||
| - | |||
| - | |||
==Protocols== | ==Protocols== | ||
| Line 45: | Line 35: | ||
[http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6WSN-418PXJ2-J&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=c7eb578c82d0073b9433d81eac64512c Crystal Structure at 1.7 Å Resolution of VEGF in Complex with Domain 2 of the Flt-1 Receptor] | [http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6WSN-418PXJ2-J&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=c7eb578c82d0073b9433d81eac64512c Crystal Structure at 1.7 Å Resolution of VEGF in Complex with Domain 2 of the Flt-1 Receptor] | ||
| - | [http://www.ncbi.nlm.nih.gov/sites/entrez/?db=gene&cmd=Retrieve&dopt=summary&list_uids=16542 flk-1 | + | [http://www.ncbi.nlm.nih.gov/sites/entrez/?db=gene&cmd=Retrieve&dopt=summary&list_uids=16542 flk-1 sequence and other information] |
[http://www.ebi.ac.uk/interpro/IEntry?ac=IPR009136 VEGFR structural info] | [http://www.ebi.ac.uk/interpro/IEntry?ac=IPR009136 VEGFR structural info] | ||
| Line 54: | Line 44: | ||
[http://www.rcsb.org/pdb/explore.do?structureId=1ZEA Structure and sequence of TE33 anti-cholera antibody] | [http://www.rcsb.org/pdb/explore.do?structureId=1ZEA Structure and sequence of TE33 anti-cholera antibody] | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=37935 TE33 antibody structure] | ||
[http://www.liebertonline.com/doi/abs/10.1089/hyb.2006.25.181?cookieSet=1&journalCode=hyb Optimizing the Generation of Recombinant Single-Chain Antibodies Against Placental Alkaline Phosphatase] | [http://www.liebertonline.com/doi/abs/10.1089/hyb.2006.25.181?cookieSet=1&journalCode=hyb Optimizing the Generation of Recombinant Single-Chain Antibodies Against Placental Alkaline Phosphatase] | ||
| Line 63: | Line 55: | ||
[http://www.springerlink.com.proxy2.library.uiuc.edu/content/b3160t3ml6430x48/ Functional coupling of the mammalian EGF receptor to the Ras/cAMP pathway in the yeast Saccharomyces cerevisiae] | [http://www.springerlink.com.proxy2.library.uiuc.edu/content/b3160t3ml6430x48/ Functional coupling of the mammalian EGF receptor to the Ras/cAMP pathway in the yeast Saccharomyces cerevisiae] | ||
| - | [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones cloned flk-1 gene] | + | [http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones cloned flk-1 gene available from ATCC] |
| + | |||
| + | ==Results== | ||
| + | *The relevant fragments of the TE33 antibody have been isolated. | ||
| + | *These fragments have been assembled into a single-chain antibody. | ||
| - | + | {{bottom_template}} | |
Latest revision as of 04:36, 30 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
Contents |
Core Team Members
Aleem, Joleen, Graham, Palak, Yanfen, Matt
Project Abstract
In the diverse world of cellular signaling pathways one of the most studied and well characterized family of receptors are the receptor tyrosine kinases. These cell surface proteins detect a wide range of extracellular ligands including hormones, growth factors, and cytokines. Receptor tyrosine kinases are most typically composed of an extracellular ligand binding domain, a single transmembrane helix, and an intracellular effector domain. Upon receptor binding of its ligand, signal cascades are activated within the cell. The first step in these signal cascades is often receptor oligomerization and cross-phosphorylation which leads to activation of MAP kinase cascades or other pathways. We plan to leverage the well understood mechanisms of receptor tyrosine kinases for the purpose of detecting the cholera toxin.
Specific Plans, Supplies, and Protocols
We have chosen yeast as our platform for this project due to it's ability to correctly express cell surface proteins. Yeast do not naturally produce receptor tyrosine kinases, but mammalian RTKs have been functionally expressed in yeast. Our first step will be to express in yeast a well understood receptor tyrosine kinase, vascular endothelial growth factor receptor in this case. From here we will create a fusion protein that will allow us to detect a ligand of our choosing. We have chosen the cholera toxin, for which an antibody against the B subunit has been sequenced. A fusion protein containing this antibody in place of VEGFR's normal ligand binding domain will bind the cholera toxin, and due to the pentameric structure of the B subunit multiple receptors will be able to dimerize and thus activate a signal cascade. Natural antibodies are composed of four separate polypeptide chains, but it is not possible to fuse all of these chains to a receptor. To get around this problem, we will create a single-chain antibody which consists of the antigen binding fragments of the light and heavy chains of an antibody joined together by a flexible glycine-serine linker. This type of antibody has been shown to retain specificity and affinity comparable to native antibodies. Finally, we will integrate the signalling event created by antigen binding and receptor dimerization into an endogenous signaling pathway, which we will then engineer to activate transcription of a reporter such as GFP.
To implement this project we will need to acquire or clone the genes for our receptor tyrosine kinase, the cholera antibody, and other signaling proteins needed to interface with the yeast pathway. Techniques such as PCR, cloning, making fusion proteins, and yeast culture will be needed. Supplies needed will include the cholera B subunit, yeast culture media, and standard molecular biology reagents/kits.
This project can be easily modularized into several subprojects. Expression of the RTK can be carried out independently of ligand binding or signalling, and this could possibly be tested by using fluorescently tagged antibodies to check for cell surface localization of the RTK. The fusion protein could be constructed and antigen binding tested perhaps by FRET or some method of affinity chromatography. The signal cascade could be tested using the unmodified RTK and detecting whether our reporter gene is activated in the presence of the RTK's natural ligand.
In Pictures
Protocols
[http://openwetware.org/wiki/Assembly_pcr Assembly PCR]
[http://openwetware.org/wiki/Knight:Colony_PCR Colony PCR]
Literature References
[http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6WSN-418PXJ2-J&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=c7eb578c82d0073b9433d81eac64512c Crystal Structure at 1.7 Å Resolution of VEGF in Complex with Domain 2 of the Flt-1 Receptor]
[http://www.ncbi.nlm.nih.gov/sites/entrez/?db=gene&cmd=Retrieve&dopt=summary&list_uids=16542 flk-1 sequence and other information]
[http://www.ebi.ac.uk/interpro/IEntry?ac=IPR009136 VEGFR structural info]
[http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6T20-4NSWYW8-3&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=6b7cee9f4b01de5cb80f1032ee714dbe Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae]
[http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6WFC-4HR76P1-2&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=b831d6adc9a66ee88647cd7baf937586 Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis]
[http://www.rcsb.org/pdb/explore.do?structureId=1ZEA Structure and sequence of TE33 anti-cholera antibody]
[http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=37935 TE33 antibody structure]
[http://www.liebertonline.com/doi/abs/10.1089/hyb.2006.25.181?cookieSet=1&journalCode=hyb Optimizing the Generation of Recombinant Single-Chain Antibodies Against Placental Alkaline Phosphatase]
[http://www.sciencedirect.com.proxy2.library.uiuc.edu/science?_ob=ArticleURL&_udi=B6T9Y-4RSS762-1&_user=571676&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000029040&_version=1&_urlVersion=0&_userid=571676&md5=d6e2fb780a92130a3e4e213502ca0448 Biodistribution and tumor imaging of an anti-CEA single-chain antibody–albumin fusion protein]
[http://www.nature.com/nbt/journal/v9/n2/abs/nbt0291-165.html Single Chain Antibody (SCA) Encoding Genes: One-Step Construction and Expression in Eukaryotic Cells]
[http://www.springerlink.com.proxy2.library.uiuc.edu/content/b3160t3ml6430x48/ Functional coupling of the mammalian EGF receptor to the Ras/cAMP pathway in the yeast Saccharomyces cerevisiae]
[http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=MGC-18600&Template=mgcMouseClones cloned flk-1 gene available from ATCC]
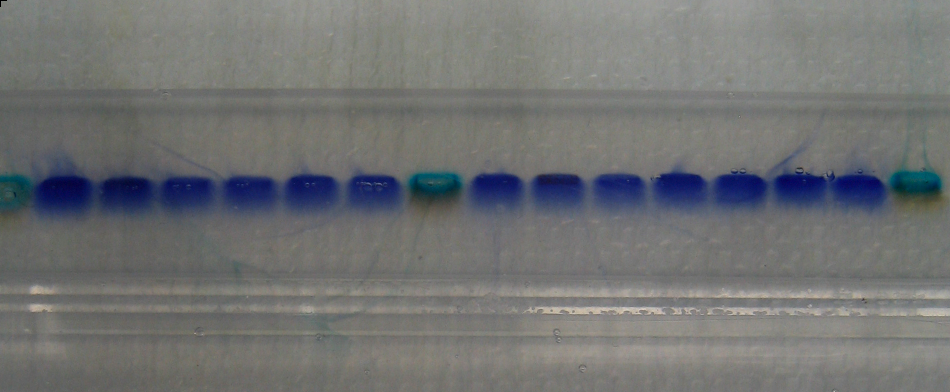
Results
- The relevant fragments of the TE33 antibody have been isolated.
- These fragments have been assembled into a single-chain antibody.
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
 "
"