Minnesota/24 June 2008
From 2008.igem.org
(Difference between revisions)
Emartin9808 (Talk | contribs) |
Emartin9808 (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 9: | Line 9: | ||
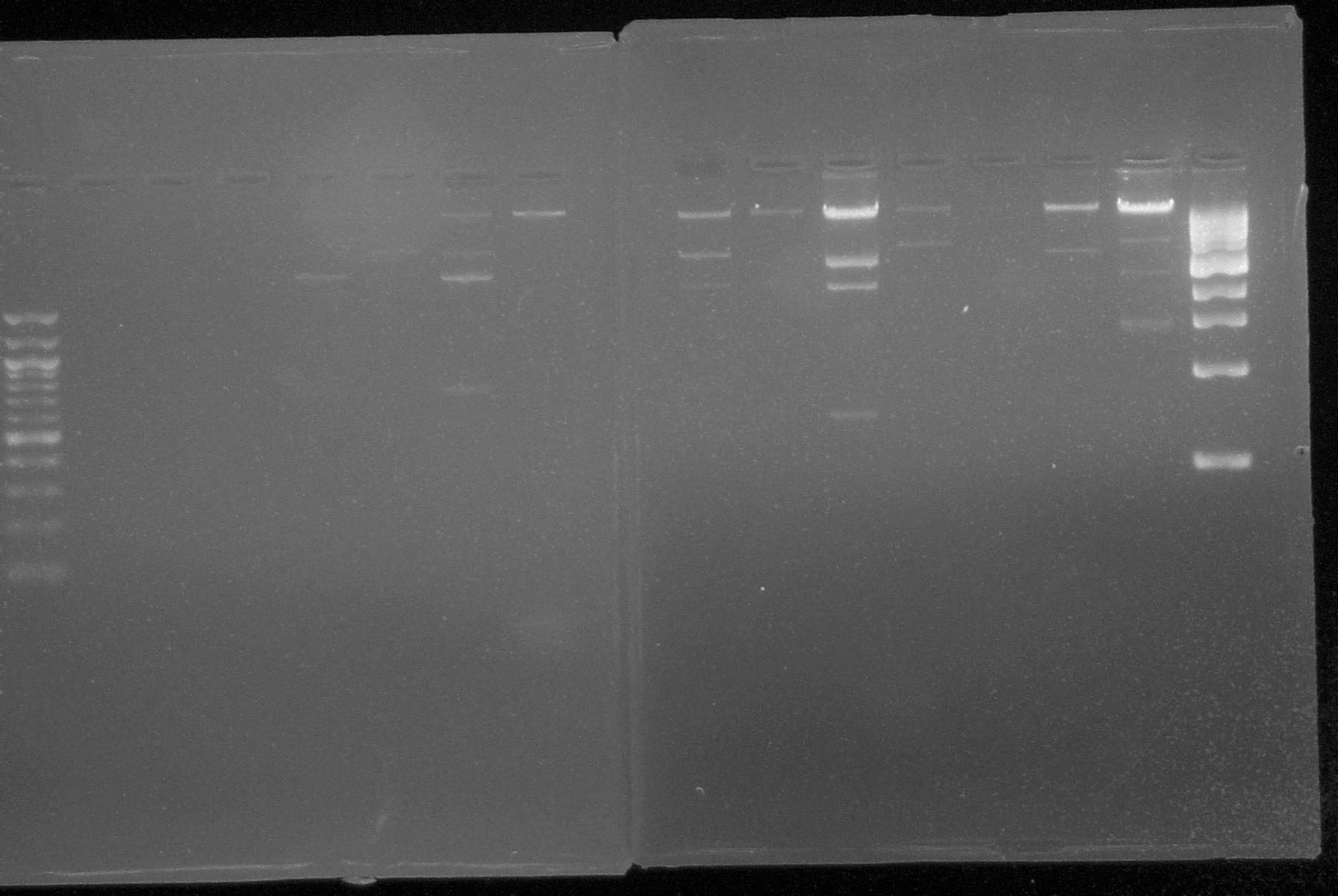
|1. '''Gel Electrophoresis:''' Used this technique to show plasmid DNA sequence. Materials: | |1. '''Gel Electrophoresis:''' Used this technique to show plasmid DNA sequence. Materials: | ||
|- | |- | ||
| - | |a. 50 uL of 1% agarose gel | + | |'''a.''' 50 uL of 1% agarose gel |
|- | |- | ||
| - | |b. TAE Buffer | + | |'''b.''' TAE Buffer |
|- | |- | ||
| - | |c. One gram of 1% agarose per 100 uL of TAE | + | |'''c.''' One gram of 1% agarose per 100 uL of TAE |
|- | |- | ||
| - | |d. Ethidium bromide (intercalating agent) | + | |'''d.''' Ethidium bromide (intercalating agent) |
|- | |- | ||
|'''Problem Encountered:''' electrophoretic gels with 1% agarose had deficient wells | |'''Problem Encountered:''' electrophoretic gels with 1% agarose had deficient wells | ||
| Line 29: | Line 29: | ||
|2. '''Plating''' from 6-23-08 transformations again. | |2. '''Plating''' from 6-23-08 transformations again. | ||
|- | |- | ||
| - | |a. Since plating of the 6-23 transformations provided no colonies for parts 15-18, the remaining cells from those transformations were re-plated. 75 uL of cell culture was spread on each of two plates for each culture; plates contained LB media and the corresponding antibiotic. A metal spreading tool was used to spread the culture suspension on the plates, and this was sterilized between each sample by dipping it in 100% ethanol (EtOH) and flaming it. 75 uL cell culture was pipetted on, and spread around plate. | + | |'''a.''' Since plating of the 6-23 transformations provided no colonies for parts 15-18, the remaining cells from those transformations were re-plated. 75 uL of cell culture was spread on each of two plates for each culture; plates contained LB media and the corresponding antibiotic. A metal spreading tool was used to spread the culture suspension on the plates, and this was sterilized between each sample by dipping it in 100% ethanol (EtOH) and flaming it. 75 uL cell culture was pipetted on, and spread around plate. |
|- | |- | ||
| - | |b. Plates were placed at 37C in an incubator and allowed to grow overnight. | + | |'''b.''' Plates were placed at 37C in an incubator and allowed to grow overnight. |
|- | |- | ||
|3. '''Sequencing primers''' ordered on 6-20-08 were picked up. All primers were diluted to mircomoles according to the following additions of sterile H20: | |3. '''Sequencing primers''' ordered on 6-20-08 were picked up. All primers were diluted to mircomoles according to the following additions of sterile H20: | ||
| + | |||
|- | |- | ||
| Line 77: | Line 78: | ||
{| | {| | ||
|- | |- | ||
| - | |a. All primers were spun down prior to opening. The appropriate amount of water was added to resuspend each primer in solutions of 100 ug/uL and 10 ug/uL. The 10 ug/uL solution is our working concentration. Primers were then stored at -20C. | + | |'''a.''' All primers were spun down prior to opening. The appropriate amount of water was added to resuspend each primer in solutions of 100 ug/uL and 10 ug/uL. The 10 ug/uL solution is our working concentration. Primers were then stored at -20C. |
|- | |- | ||
| - | |b. 12 uL reactions containing plasmid to be sequenced and the corresponding primer(s) were set up and submitted for sequencing. Each reaction mixture contained 1 ul primer, 1 ul plasmid DNA, and 10 uL dd H2O. All samples were sent for sequencing at the Biomedical Genomics Center at the U of M. | + | |'''b.''' 12 uL reactions containing plasmid to be sequenced and the corresponding primer(s) were set up and submitted for sequencing. Each reaction mixture contained 1 ul primer, 1 ul plasmid DNA, and 10 uL dd H2O. All samples were sent for sequencing at the Biomedical Genomics Center at the U of M. |
|} | |} | ||
Latest revision as of 21:17, 8 July 2008
| Back to Notebook Home | |
| Go to Previous Day (June 23) | Go to Next Day (June 25) |
| 1. Gel Electrophoresis: Used this technique to show plasmid DNA sequence. Materials: |
| a. 50 uL of 1% agarose gel |
| b. TAE Buffer |
| c. One gram of 1% agarose per 100 uL of TAE |
| d. Ethidium bromide (intercalating agent) |
| Problem Encountered: electrophoretic gels with 1% agarose had deficient wells |
| Solution: add 0.5 grams more of agarose to the 100uL of TAE buffer |
| 2. Plating from 6-23-08 transformations again. | ||
| a. Since plating of the 6-23 transformations provided no colonies for parts 15-18, the remaining cells from those transformations were re-plated. 75 uL of cell culture was spread on each of two plates for each culture; plates contained LB media and the corresponding antibiotic. A metal spreading tool was used to spread the culture suspension on the plates, and this was sterilized between each sample by dipping it in 100% ethanol (EtOH) and flaming it. 75 uL cell culture was pipetted on, and spread around plate. | ||
| b. Plates were placed at 37C in an incubator and allowed to grow overnight. | ||
| 3. Sequencing primers ordered on 6-20-08 were picked up. All primers were diluted to mircomoles according to the following additions of sterile H20: |
| Primer | nmoles | uL H20 added |
|---|---|---|
| P22 cII cR | 37.7 | 377.0 |
| P22 cII cF | 36.0 | 360.0 |
| Lambda cI R | 32.40 | 324.0 |
| Lambda cI F | 33.2 | 332.0 |
| P22 MNT R | 32.8 | 328.0 |
| P22 MNT F | 28.2 | 282.0 |
| EYFP R | 43.8 | 438.0 |
| EYFP F | 34.5 | 345.0 |
| pSB 2K3 | 39.6 | 396.0 |
| pSB 1A2 | 31.7 | 317.0 |
| pSB 1AK3 | 40.0 | 400.0 |
| GFP R | 32.1 | 321.0 |
| GFP F | 33.7 | 337.0 |
| mCherry R | 43.9 | 439.0 |
| mCherry F | 28.4 | 284.0 |
| LacI R | 29.4 | 294.0 |
| LacI F | 31.2 | 312.0 |
| a. All primers were spun down prior to opening. The appropriate amount of water was added to resuspend each primer in solutions of 100 ug/uL and 10 ug/uL. The 10 ug/uL solution is our working concentration. Primers were then stored at -20C. |
| b. 12 uL reactions containing plasmid to be sequenced and the corresponding primer(s) were set up and submitted for sequencing. Each reaction mixture contained 1 ul primer, 1 ul plasmid DNA, and 10 uL dd H2O. All samples were sent for sequencing at the Biomedical Genomics Center at the U of M. |
 "
"