Team:University of Lethbridge/Notebook/GeneralLabJuly
From 2008.igem.org
Munima.alam (Talk | contribs) m |
Munima.alam (Talk | contribs) m |
||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Team:University_of_Lethbridge/Notebook|Back to The University of Lethbridge Main Notebook]] | ||
| + | |||
===July 1, 2008=== | ===July 1, 2008=== | ||
====Nathan Phillips, Andrew==== | ====Nathan Phillips, Andrew==== | ||
Made 500 mL of LB agar + amp and 500 mL of Liquid LB | Made 500 mL of LB agar + amp and 500 mL of Liquid LB | ||
| + | |||
===July 2, 2008=== | ===July 2, 2008=== | ||
| Line 23: | Line 26: | ||
Stored in the iGEM 4 C fridge | Stored in the iGEM 4 C fridge | ||
| - | ====Nathan Puhl, Alix==== | + | ====Nathan Puhl, Alix, Sebastian==== |
Flourescent Reporter | Flourescent Reporter | ||
| Line 36: | Line 39: | ||
-centrifuge 3 min. | -centrifuge 3 min. | ||
-2 uL of plasmid into 25 uL DH5alpha | -2 uL of plasmid into 25 uL DH5alpha | ||
| - | -leave on ice for 30 min. (Left remaining DT, TetR, LacI to sit overnight at room temp. | + | -leave on ice for 30 min. |
| + | -(Left remaining DT, TetR, LacI to sit overnight at room temp. testing for better DNA recovery) | ||
-put in 42 C water bath for 45 sec. | -put in 42 C water bath for 45 sec. | ||
-chill on ice for 2 min. | -chill on ice for 2 min. | ||
| Line 49: | Line 53: | ||
-pLACI (BBa_R0011) | -pLACI (BBa_R0011) | ||
-pSTRONG (BBa_J23119) | -pSTRONG (BBa_J23119) | ||
| + | |||
| + | loading dye [http://openwetware.org/wiki/Agarose_gel_loading_dye sizes] | ||
| + | |||
| + | |||
| + | ===July 14, 2008=== | ||
| + | ====Munima, Nathan Puhl, Alix, Andrew==== | ||
| + | Ran gel electrophoresis on "comp RP1616 + pSB1A7" cells, LacI, TetR and DT (from July 8/08). | ||
| + | No DNA bands were observed. | ||
| + | |||
| + | |||
| + | ===July 15, 2008=== | ||
| + | ====Nathan Puhl, Andrew, Alix==== | ||
| + | Objective: Working on fluorescent reporter system; trying transformation from BioBricks again. | ||
| + | Using: LacI (BBa_J24679) and DT (BBa_B0015) | ||
| + | |||
| + | Protocol - "Transformation - F00 v.1" (Glinko Bioworks): | ||
| + | |||
| + | 1. Get DNA from filter paper. | ||
| + | 2. Place punch directly in 25 uL DH5alpha cells. | ||
| + | 3. Leave on ice for 30 mins. | ||
| + | 4. 45 second heat shock in 42 C water bath. | ||
| + | 5. Chill on ice for 2 minutes. | ||
| + | 6. Add 500 uL SOC broth. | ||
| + | 7. Incubate at 37 C for 1 hour. | ||
| + | 8. Spin down all cells. | ||
| + | 9. Remove 400 uL. | ||
| + | 10. Resuspend and plate on LB + Amp. Allow to incubate at 37 C overnight. | ||
| + | |||
| + | |||
| + | ===July 16, 2008=== | ||
| + | ====Andrew, Alix==== | ||
| + | Objective: Assess transformation success | ||
| + | - observed growth on LacI and DT plates | ||
| + | - grew up cells with LB + Amp | ||
| + | - put into shaker incubator at 37 C overnight | ||
| + | |||
| + | |||
| + | ===July 17, 2008=== | ||
| + | ====Nathan Puhl, Alix, Munima, Christa==== | ||
| + | Objective: Assess transformation success and isolate a single colony for subculture | ||
| + | - Streaked colonies onto Brent's LB + Amp plates because there was too much growth on the LacI/DT plates | ||
| + | |||
| + | ====Munima, Christa==== | ||
| + | Made 500mL of LB + amp semi-solid media and poured 25 plates. | ||
| + | Stored in the iGEM 4 C fridge | ||
| + | |||
| + | |||
| + | ===July 21, 2008=== | ||
| + | ====Nathan Puhl==== | ||
| + | Objective: Subculture transformed cells for plasmid isolation | ||
| + | - Subcultured single colony of re-streaked transformants in LB + amp and shaker incubated overnight at 37 C | ||
| + | |||
| + | |||
| + | ===July 22, 2008=== | ||
| + | ====Nathan Puhl, Andrew==== | ||
| + | Objective:Isolate plasmid from subcultured transformed cells and attempt to transform three more biobricks from the filter paper into ''E.coli'' | ||
| + | - Plasmid prepped BBa_J24679 (LacI) and BBa_B0015 (DT) from transformed DH5-alpha | ||
| + | - Transformed DH5-alpha with BBa_P0440 (TetR), BBa_I714062 (GTPTT), and BBa_J31007 (TetA) as per the protocol from July 15, 2008 | ||
| + | |||
| + | Objective: Optimize PCR conditions for newly received primers. | ||
| + | |||
| + | Set up PCR for CheZ and Riboswitch. | ||
| + | -Used 1/10, 1/100, 1/1000 dilutions of pTopp for template | ||
| + | |||
| + | PCR conditions: | ||
| + | A. Initial denaturation: 98 C (30 sec) | ||
| + | B. -Denaturation: 98 C (10 sec) | ||
| + | -Annealing: 50 C (15 sec) | ||
| + | -Extension 72 C (30 sec) | ||
| + | -35 cycles of step B. | ||
| + | C.Final extension: 72 C (7 min); 4 C (inifinity) | ||
| + | |||
| + | |||
| + | ===July 25, 2008=== | ||
| + | ====Christa==== | ||
| + | Did a Eppendorf FastPlasmid Mini-prep on cells transformed with Tet A. Stored in -20 C labelled as Tet A (1 or 2) July 25/08 | ||
| + | |||
| + | |||
| + | ===July 28, 2008=== | ||
| + | ====Nathan Puhl, Alix==== | ||
| + | Objective: Riboswitch PCR (under Riboswitch: July notebook) | ||
| + | |||
| + | |||
| + | ===July 29, 2008=== | ||
| + | ====Nathan Puhl, Roxanne, Alix==== | ||
| + | Streak-Plated cells from the 6 Stab Culture Tubes received from iGEM to determine if the cells are viable. | ||
| + | Incubated overnight. | ||
| + | |||
| + | BBa_I13401, BBa_I13504, BBa_C0012 (LacI), BBa_P0440 (TetR), BBa_B0015 (DT), BBa_J13007 (TetA) | ||
| + | |||
| + | |||
| + | ===July 30, 2008=== | ||
| + | ====Roxanne, Nathan Puhl==== | ||
| + | Picked Colonies from the Biobrick Plates and Incubated them in LB Media overnight. | ||
| + | |||
| + | ====Roxanne==== | ||
| + | Shadowed John (for his own work) for a complete day of cloning (Restriction digest, Ligation, Transformation and Plating). | ||
| + | |||
| + | |||
| + | ===July 31, 2008=== | ||
| + | ====Roxanne, Jaden==== | ||
| + | -Made Glycerol stocks of the 6 Biobrick parts. | ||
| + | -Plasmid Prepped the 6 Biobrick Parts using a Qiagen miniprep kit. | ||
| + | |||
| + | ====Roxanne==== | ||
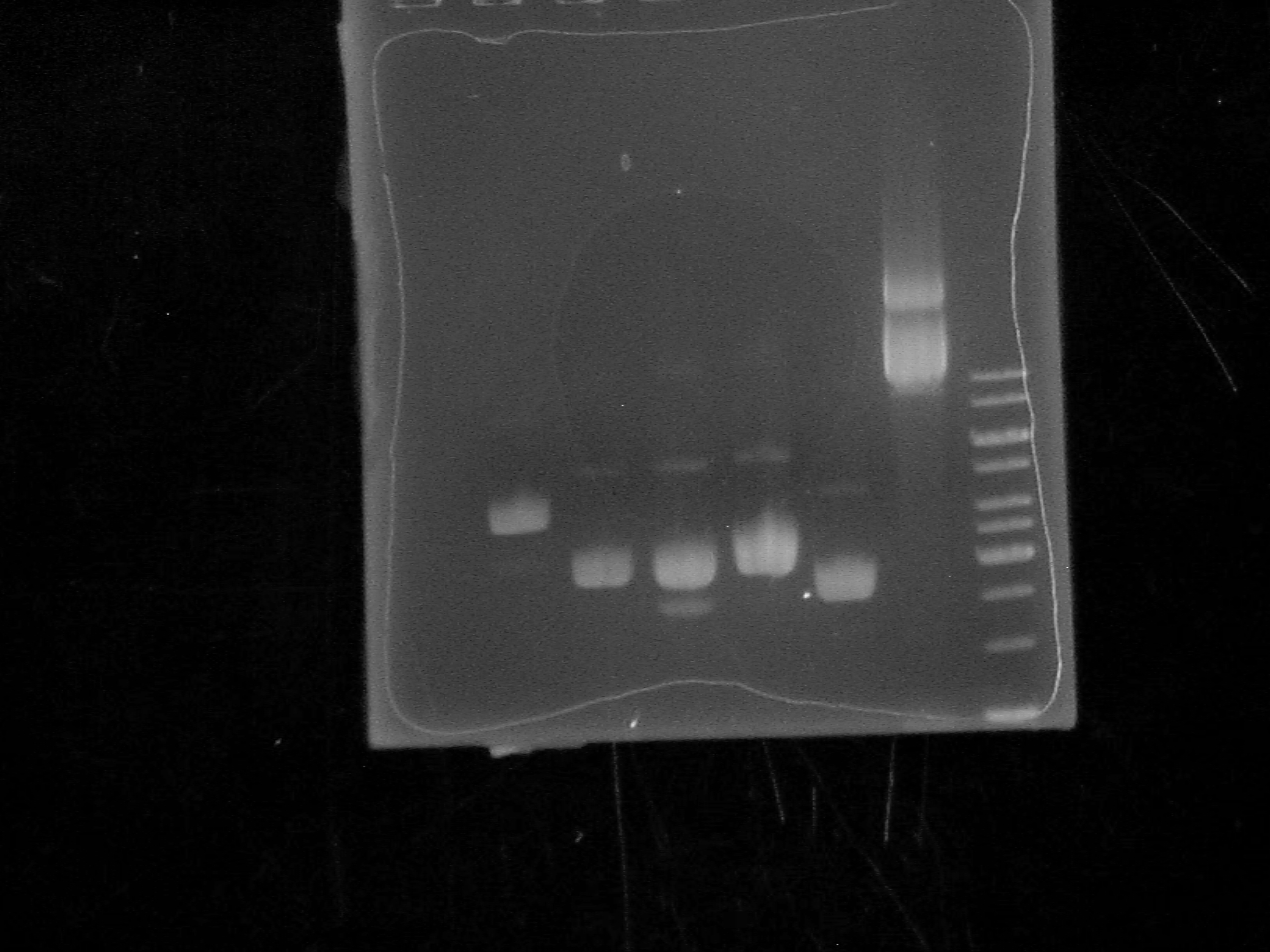
| + | -Ran a 1% Agarose Gel of the 6 Biobrick plasmids at 120V for 34 minutes. | ||
| + | |||
| + | |||
| + | [[Image: Biobrick_Gel.jpg|500 px]] | ||
| + | |||
| + | |||
| + | Lane 1: empty | ||
| + | |||
| + | Lane 2: BBa_I13401 | ||
| + | |||
| + | Lane 3: BBa_C0012 | ||
| + | |||
| + | Lane 4: BBa_J31007 | ||
| + | |||
| + | Lane 5: BBa_B0015 | ||
| + | |||
| + | Lane 6: BBa_P0440 | ||
| + | |||
| + | Lane 7: BBa_I13504 | ||
| + | |||
| + | Lane 8: Ladder | ||
Latest revision as of 02:29, 30 October 2008
Back to The University of Lethbridge Main Notebook
Contents |
July 1, 2008
Nathan Phillips, Andrew
Made 500 mL of LB agar + amp and 500 mL of Liquid LB
July 2, 2008
Nathan Puhl, Alix, Sebastian, Munima, Roxanne, Christa
Transformed [http://partsregistry.org/wiki/index.php?title=Part:BBa_J24679 BBa_J24679] (RBS + LacI), [http://partsregistry.org/wiki/index.php?title=Part:BBa_P0440 BBa_P0440] (RBS+TetR+T10+T12), leftover DNA from Double T (June 26, 2008), and 1 uL of pSB1A7 plasmid (June 18, 2008).
Protocol changes:
-3 uL of DNA -spin down 200 uL of cells and resuspend in 100 uL of LB; plate on LB + amp
July 3, 2008
Nathan Puhl, Munima, Christa, Alix, Roxanne, Sebastian
Checked transformation plates. Only the positive control (pSB1A7) had colonies (~1500) indicating that there is nothing wrong with the transformation protocol or cells so we must be having problems with the DNA extraction from the filter paper. Next week we will attempt various changes to the protocol to extract more DNA.
July 8, 2008
Munima, Christa, Nathan Puhl
Made 500mL of LB semi-solid media and poured 24 plates. Stored in the iGEM 4 C fridge
Nathan Puhl, Alix, Sebastian
Flourescent Reporter
Transformation from BioBricks LacI (BBA_J24679), TetR (BBa_P0440) and DT (BBa_B0015).
Protocol:
-punched out 2 spots of filter paper
-15uL TE, for 30 min. @ 50 C
-centrifuge 3 min. @ 15000 g
-freeze 5 min. in -20 C freezer
-heat shock 1 min. in 42 C water bath
-centrifuge 3 min.
-2 uL of plasmid into 25 uL DH5alpha
-leave on ice for 30 min.
-(Left remaining DT, TetR, LacI to sit overnight at room temp. testing for better DNA recovery)
-put in 42 C water bath for 45 sec.
-chill on ice for 2 min.
-add 1 mL of SOC broth
-incubate cells @ 37 C, 225 rpm for 60 min.
-spin down 400 uL cells (1 min. 16000xG), remove 300 uL
-resuspend
-plate, incubate at 37 C overnight
Subcultured 3 biobricks (for flourescent reporter) glycerol stocked from last year into 5 mL liquid LB + Amp.
-RFP Sub. (BBa_I13507) -pLACI (BBa_R0011) -pSTRONG (BBa_J23119)
loading dye [http://openwetware.org/wiki/Agarose_gel_loading_dye sizes]
July 14, 2008
Munima, Nathan Puhl, Alix, Andrew
Ran gel electrophoresis on "comp RP1616 + pSB1A7" cells, LacI, TetR and DT (from July 8/08). No DNA bands were observed.
July 15, 2008
Nathan Puhl, Andrew, Alix
Objective: Working on fluorescent reporter system; trying transformation from BioBricks again. Using: LacI (BBa_J24679) and DT (BBa_B0015)
Protocol - "Transformation - F00 v.1" (Glinko Bioworks):
1. Get DNA from filter paper. 2. Place punch directly in 25 uL DH5alpha cells. 3. Leave on ice for 30 mins. 4. 45 second heat shock in 42 C water bath. 5. Chill on ice for 2 minutes. 6. Add 500 uL SOC broth. 7. Incubate at 37 C for 1 hour. 8. Spin down all cells. 9. Remove 400 uL. 10. Resuspend and plate on LB + Amp. Allow to incubate at 37 C overnight.
July 16, 2008
Andrew, Alix
Objective: Assess transformation success
- observed growth on LacI and DT plates - grew up cells with LB + Amp - put into shaker incubator at 37 C overnight
July 17, 2008
Nathan Puhl, Alix, Munima, Christa
Objective: Assess transformation success and isolate a single colony for subculture
- Streaked colonies onto Brent's LB + Amp plates because there was too much growth on the LacI/DT plates
Munima, Christa
Made 500mL of LB + amp semi-solid media and poured 25 plates. Stored in the iGEM 4 C fridge
July 21, 2008
Nathan Puhl
Objective: Subculture transformed cells for plasmid isolation
- Subcultured single colony of re-streaked transformants in LB + amp and shaker incubated overnight at 37 C
July 22, 2008
Nathan Puhl, Andrew
Objective:Isolate plasmid from subcultured transformed cells and attempt to transform three more biobricks from the filter paper into E.coli
- Plasmid prepped BBa_J24679 (LacI) and BBa_B0015 (DT) from transformed DH5-alpha - Transformed DH5-alpha with BBa_P0440 (TetR), BBa_I714062 (GTPTT), and BBa_J31007 (TetA) as per the protocol from July 15, 2008
Objective: Optimize PCR conditions for newly received primers.
Set up PCR for CheZ and Riboswitch.
-Used 1/10, 1/100, 1/1000 dilutions of pTopp for template
PCR conditions:
A. Initial denaturation: 98 C (30 sec)
B. -Denaturation: 98 C (10 sec)
-Annealing: 50 C (15 sec)
-Extension 72 C (30 sec)
-35 cycles of step B.
C.Final extension: 72 C (7 min); 4 C (inifinity)
July 25, 2008
Christa
Did a Eppendorf FastPlasmid Mini-prep on cells transformed with Tet A. Stored in -20 C labelled as Tet A (1 or 2) July 25/08
July 28, 2008
Nathan Puhl, Alix
Objective: Riboswitch PCR (under Riboswitch: July notebook)
July 29, 2008
Nathan Puhl, Roxanne, Alix
Streak-Plated cells from the 6 Stab Culture Tubes received from iGEM to determine if the cells are viable. Incubated overnight.
BBa_I13401, BBa_I13504, BBa_C0012 (LacI), BBa_P0440 (TetR), BBa_B0015 (DT), BBa_J13007 (TetA)
July 30, 2008
Roxanne, Nathan Puhl
Picked Colonies from the Biobrick Plates and Incubated them in LB Media overnight.
Roxanne
Shadowed John (for his own work) for a complete day of cloning (Restriction digest, Ligation, Transformation and Plating).
July 31, 2008
Roxanne, Jaden
-Made Glycerol stocks of the 6 Biobrick parts. -Plasmid Prepped the 6 Biobrick Parts using a Qiagen miniprep kit.
Roxanne
-Ran a 1% Agarose Gel of the 6 Biobrick plasmids at 120V for 34 minutes.
Lane 1: empty
Lane 2: BBa_I13401
Lane 3: BBa_C0012
Lane 4: BBa_J31007
Lane 5: BBa_B0015
Lane 6: BBa_P0440
Lane 7: BBa_I13504
Lane 8: Ladder
 "
"