|
|
| (50 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Paris/Menu}} | | {{Paris/Menu}} |
| | | | |
| - | <center> <FONT size=5> '''All the parameters we might have to introduce in our models''' </FONT> <BR> </center> | + | <center> <FONT size=5> '''References''' </FONT> <BR> </center> |
| | | | |
| - | == <math> flhDC \rightarrow fliA </math> ==
| + | <br> |
| - | <FONT size=6> flhDC → fliA </FONT> <BR>
| + | * [1] Shiraz Kalir, Uri Alon. Using quantitative blueprint to reprogram the dynamics of the flagella network. Cell, June 11, 2004, Vol.117, 713-720. |
| | + | * [2] Jordi Garcia-Ojalvo, Michael B. Elowitz, Steven H. Strogratz. Modeling a synthetic multicellular clock : repressilators coupled by quorum sensing. PNAS, July 27, 204, Vol. 101, no. 30. |
| | + | * [3] Nitzan Rosenfeld, Uri Alon. Response delays and the structure of transcription networks. JMB, 2003, 329, 645-654. |
| | + | * [4] Nitzan Rosenfeld, Michael B. Elowitz, Uri Alon. Negative autoregulation speeds the response times of transcription networks. JMB, 2003, 323, 785-793. |
| | + | * [5] S.Kalir, J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, U. Alon. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science, June 2001, Vol 292. |
| | + | * [6] http://partsregistry.org/Part:BBa_E0040 |
| | + | * [7] M. Schuster, M. L. Urbanowski, and E. P. Greenberg. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. |
| | + | * [8] Sheng Jian Cai and Masayori Inouye. EnvZ-OmpR Interaction and Osmoregulation in ''Escherichia coli''. The journal of biochemical chemistry Vol. 277, No. 27, Issue of July 5, pp. 24155–24161, 2002. |
| | | | |
| - | == <math> flhDC \rightarrow fliL </math> ==
| |
| | | | |
| - | == <math> flhDC \rightarrow flgA</math> ==
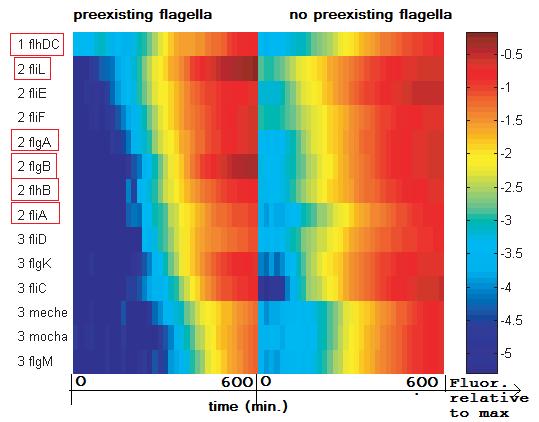
| + | <center>[[Image:Diag_expr_time.jpg]]</center> |
| - | | + | <center> ''expression of the fl-genes observed in'' [[Team:Paris/Modeling/Bibliography|[5]]] </center> |
| - | == <math> flhDC \rightarrow flgB </math> ==
| + | |
| - | | + | |
| - | == <math> flhDC \rightarrow flhB </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow fliA </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow fliL </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flgA </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flgB </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flhB </math> ==
| + | |
| - | | + | |
| - | == <math> flhDC \rightarrow \emptyset \</math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow \emptyset </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flgA </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flgB </math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow flhB </math> ==
| + | |
| - | | + | |
| - | == <math> flhDC \rightarrow \emptyset \</math> ==
| + | |
| - | | + | |
| - | == <math> fliA \rightarrow \emptyset </math> ==
| + | |
| - | | + | |
| - | == <math> fliL \rightarrow \emptyset </math> ==
| + | |
| - | | + | |
| - | == <math> flgA \rightarrow \emptyset </math> ==
| + | |
| - | | + | |
| - | == <math> flgB \rightarrow \emptyset \</math> ==
| + | |
| - | | + | |
| - | == <math> flhB \rightarrow \emptyset </math> ==
| + | |
| - | | + | |
| - | == <math> lasI \rightarrow 3OC12HSL </math> ==
| + | |
| - | | + | |
| - | == <math> \frac{\partial HSL}{\partial t} = D*\frac{\partial ^{2} HSL}{\partial x} ==
| + | |
References
- [1] Shiraz Kalir, Uri Alon. Using quantitative blueprint to reprogram the dynamics of the flagella network. Cell, June 11, 2004, Vol.117, 713-720.
- [2] Jordi Garcia-Ojalvo, Michael B. Elowitz, Steven H. Strogratz. Modeling a synthetic multicellular clock : repressilators coupled by quorum sensing. PNAS, July 27, 204, Vol. 101, no. 30.
- [3] Nitzan Rosenfeld, Uri Alon. Response delays and the structure of transcription networks. JMB, 2003, 329, 645-654.
- [4] Nitzan Rosenfeld, Michael B. Elowitz, Uri Alon. Negative autoregulation speeds the response times of transcription networks. JMB, 2003, 323, 785-793.
- [5] S.Kalir, J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, U. Alon. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science, June 2001, Vol 292.
- [6] http://partsregistry.org/Part:BBa_E0040
- [7] M. Schuster, M. L. Urbanowski, and E. P. Greenberg. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR.
- [8] Sheng Jian Cai and Masayori Inouye. EnvZ-OmpR Interaction and Osmoregulation in Escherichia coli. The journal of biochemical chemistry Vol. 277, No. 27, Issue of July 5, pp. 24155–24161, 2002.
 expression of the fl-genes observed in [5]
expression of the fl-genes observed in [5]
|
 "
"

