Team:KULeuven/24 July 2008
From 2008.igem.org
(→Wet Lab) |
m |
||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Header}} | ||
| + | |||
{{:Team:KULeuven/Tools/New_Day/Date_Retriever}} | {{:Team:KULeuven/Tools/New_Day/Date_Retriever}} | ||
| Line 5: | Line 8: | ||
=== Wet Lab === | === Wet Lab === | ||
| - | Stefanie and Hanne made a | + | Stefanie and Hanne made a glycerol stock of all the parts we received from the iGEM headquarters. Everything is now stored in two -80°C freezers. You can find the cells with the parts in box S&P-F7. You can look up in the cmpg database where you should search for a specific part. |
| - | + | ||
| + | In the afternoon we started the protocol to make the desired ''E.coli'' strain by transducing the EnvZ-mutation from V1012 to MC1061 cells. Therefore we made a liquid culture of the donor and acceptor cells. Phage P1 was incubated with the acceptor. Tomorrow we'll continue the proces and bring phage P1 and the acceptor cells (MC1061) together, this should result in the right cells. | ||
| - | Because the concentrations of the isolated plasmids were low yesterday, we | + | Because the concentrations of the isolated plasmids were low yesterday, Elke, Jan, Andim and Nathalie did it all over again. However, this time we tried to obtain higher concentrations of plasmid DNA. Normally, this will improve the success rate of the digests. We did this for the following BioBricks: |
| - | + | {| style="background:#99CCFF;color:black;width:60%;margin-left:50px;" border="1" cellpadding="2" cellspacing="0" | |
| - | + | ! BioBrick !! Sample 1: Concentration !! Sample 2: Concentration | |
| - | + | |- style="background:#ffffff; color:black; text-align: center;" | |
| - | + | | [http://partsregistry.org/Part:BBa_B0032 B0032] || 114.8 ng/µl || / | |
| - | + | |- style="background:#ffffff; color:black; text-align: center;" | |
| - | + | | [http://partsregistry.org/Part:BBa_B0034 B0034] || 73.8 ng/µl || 95.1 ng/µl | |
| - | + | |- style="background:#ffffff; color:black; text-align: center;" | |
| - | + | | [http://partsregistry.org/Part:BBa_E0022 E0022] || 86.4 ng/µl || / | |
| - | Then the plasmids were digested with the restriction enzymes (''Eco'' | + | |- style="background:#ffffff; color:black; text-align: center;" |
| + | | [http://partsregistry.org/Part:BBa_I714891 I714891] || 140.9 ng/µl || / | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_J23100 J23100] || 345.0 ng/µl || 176.1 ng/µl | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_M30109 M30109] || 54.3 ng/µl || / | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_R0084 R0084] || 66.9 ng/µl || 60.9 ng/µl | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_B0015 B0015] || 112.3 ng/µl || 130.3 ng/µl | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_C0040 C0040] || 177.1 ng/µl || 161.7 ng/µl | ||
| + | |} | ||
| + | <br> | ||
| + | |||
| + | Then the plasmids were digested with the appropriate restriction enzymes (''Eco''R I, ''Spe'' I, ''Xba'' I). In order to see whether the digestion worked, we put the samples on an agarose gel. The plasmids with BioBricks [http://partsregistry.org/Part:BBa_I714891 I714891] and [http://partsregistry.org/Part:BBa_B0015 B0015] were not cut properly by the enzymes. The other parts were sliced out of the gel and are ready to be purified and ligated. | ||
| + | {| style="background:#99CCFF;color:black;width:60%;margin-left:50px;" border="1" cellpadding="2" cellspacing="0" | ||
| + | ! BioBrick !! Restriction enzymes !! length desired fragment !! success rate digestion | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_B0032 B0032] || ''Eco''R I, ''Xba'' I || 2077 || probably | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_B0034 B0034] || ''Eco''R I, ''Xba'' I || 2077 || probably | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_E0022 E0022] || ''Eco''R I, ''Spe'' I || 783 || OK | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_I714891 I714891] || ''Eco''R I, ''Spe'' I || 757 || failed | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_J23100 J23100] || ''Eco''R I, ''Spe'' I || 51 || OK | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_R0084 R0084] || ''Eco''R I, ''Spe'' I || 129 || OK | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_B0015 B0015] || ''Eco''R I, ''Xba'' I || 2193 || failed | ||
| + | |- style="background:#ffffff; color:black; text-align: center;" | ||
| + | | [http://partsregistry.org/Part:BBa_C0040 C0040] || ''Eco''R I, ''Spe'' I || 681 || OK | ||
| + | |} | ||
| + | <br> | ||
=== Dry Lab === | === Dry Lab === | ||
| + | |||
| + | ==== Primers ==== | ||
Primers for the construction of T7 polymerase with UmuD tag were written out. | Primers for the construction of T7 polymerase with UmuD tag were written out. | ||
| - | == Modeling == | + | ==== Modeling ==== |
| - | + | Pulse generator has been reviewed, the parameters have been checked but there are issues... The pulse generator doesn't work (pure physically): when a step signal of T7 is applied cI is produced, which will then repress its transcription. Of course, this will develop to a steady state with a cI level equal to a non zero value. A zero cI value would result in zero repression, thereby restarting the transcription of the cI gene and start producing cI again anyway. Because the cI value remains non zero, so will the lactonase production, and so there will always be a constant amount of lactonase present. Question is: how much will this concentration affect the system considering the case of no light present? No light present means HSL production, which should bind to LuxR, but instead will react with lactonase to an hydroxy-acid. If the concentration of lactonase is considerable, and remains somewhat steady, cell death would never occur. Interesting link to a pulse generator system that seems to work: | |
| - | Pulse generator has been reviewed, the parameters have been checked but there are issues... | + | [https://keaslinglab.lbl.gov/wiki/index.php/Synthetic_Biology_-_Devices_-_Pulse_Generator Pulse Generator Keasling Lab] |
| - | + | {| | |
| - | A zero cI value would result in zero repression, thereby restarting the transcription of the cI gene and start producing cI again anyway. | + | |- |
| - | + | |[[image:Sim_pulse_generator_Lac.jpg|left|420px]]||[[Image:Sim_pulse_generator_cI.jpg|right|420px]] | |
| - | Because the cI value remains non zero, so will the lactonase production, and so there will always be a constant amount of lactonase present. Question is how much this concentration | + | |
| - | + | ||
| - | {| | + | |
| - | |[[ | + | |
| - | |[[Image:Sim_pulse_generator_cI.jpg| | + | |
|} | |} | ||
| - | + | The lactonase amount does receed to 0 when the filter has 0 output, when the inverter gives high input, but this could cause delay to cell death, due to the slow degradation of lactonase. | |
| - | [ | + | [[Image:Sim_pulse_generator_pulses.jpg|420px|center]] |
| + | <br> | ||
The memory system has been reviewed as well. The old system seemed unsuccesful, a new system has been found proven to work. There are however not enough repressors available to implement this system without having interference with other parts. ... | The memory system has been reviewed as well. The old system seemed unsuccesful, a new system has been found proven to work. There are however not enough repressors available to implement this system without having interference with other parts. ... | ||
| Line 47: | Line 83: | ||
== Quote of the day == | == Quote of the day == | ||
| - | |||
GAS! | GAS! | ||
Latest revision as of 13:07, 7 October 2008
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
Contents |
Lab Work
Wet Lab
Stefanie and Hanne made a glycerol stock of all the parts we received from the iGEM headquarters. Everything is now stored in two -80°C freezers. You can find the cells with the parts in box S&P-F7. You can look up in the cmpg database where you should search for a specific part.
In the afternoon we started the protocol to make the desired E.coli strain by transducing the EnvZ-mutation from V1012 to MC1061 cells. Therefore we made a liquid culture of the donor and acceptor cells. Phage P1 was incubated with the acceptor. Tomorrow we'll continue the proces and bring phage P1 and the acceptor cells (MC1061) together, this should result in the right cells.
Because the concentrations of the isolated plasmids were low yesterday, Elke, Jan, Andim and Nathalie did it all over again. However, this time we tried to obtain higher concentrations of plasmid DNA. Normally, this will improve the success rate of the digests. We did this for the following BioBricks:
| BioBrick | Sample 1: Concentration | Sample 2: Concentration |
|---|---|---|
| [http://partsregistry.org/Part:BBa_B0032 B0032] | 114.8 ng/µl | / |
| [http://partsregistry.org/Part:BBa_B0034 B0034] | 73.8 ng/µl | 95.1 ng/µl |
| [http://partsregistry.org/Part:BBa_E0022 E0022] | 86.4 ng/µl | / |
| [http://partsregistry.org/Part:BBa_I714891 I714891] | 140.9 ng/µl | / |
| [http://partsregistry.org/Part:BBa_J23100 J23100] | 345.0 ng/µl | 176.1 ng/µl |
| [http://partsregistry.org/Part:BBa_M30109 M30109] | 54.3 ng/µl | / |
| [http://partsregistry.org/Part:BBa_R0084 R0084] | 66.9 ng/µl | 60.9 ng/µl |
| [http://partsregistry.org/Part:BBa_B0015 B0015] | 112.3 ng/µl | 130.3 ng/µl |
| [http://partsregistry.org/Part:BBa_C0040 C0040] | 177.1 ng/µl | 161.7 ng/µl |
Then the plasmids were digested with the appropriate restriction enzymes (EcoR I, Spe I, Xba I). In order to see whether the digestion worked, we put the samples on an agarose gel. The plasmids with BioBricks [http://partsregistry.org/Part:BBa_I714891 I714891] and [http://partsregistry.org/Part:BBa_B0015 B0015] were not cut properly by the enzymes. The other parts were sliced out of the gel and are ready to be purified and ligated.
| BioBrick | Restriction enzymes | length desired fragment | success rate digestion |
|---|---|---|---|
| [http://partsregistry.org/Part:BBa_B0032 B0032] | EcoR I, Xba I | 2077 | probably |
| [http://partsregistry.org/Part:BBa_B0034 B0034] | EcoR I, Xba I | 2077 | probably |
| [http://partsregistry.org/Part:BBa_E0022 E0022] | EcoR I, Spe I | 783 | OK |
| [http://partsregistry.org/Part:BBa_I714891 I714891] | EcoR I, Spe I | 757 | failed |
| [http://partsregistry.org/Part:BBa_J23100 J23100] | EcoR I, Spe I | 51 | OK |
| [http://partsregistry.org/Part:BBa_R0084 R0084] | EcoR I, Spe I | 129 | OK |
| [http://partsregistry.org/Part:BBa_B0015 B0015] | EcoR I, Xba I | 2193 | failed |
| [http://partsregistry.org/Part:BBa_C0040 C0040] | EcoR I, Spe I | 681 | OK |
Dry Lab
Primers
Primers for the construction of T7 polymerase with UmuD tag were written out.
Modeling
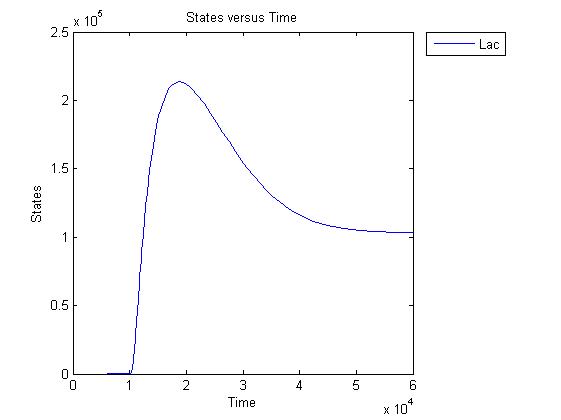
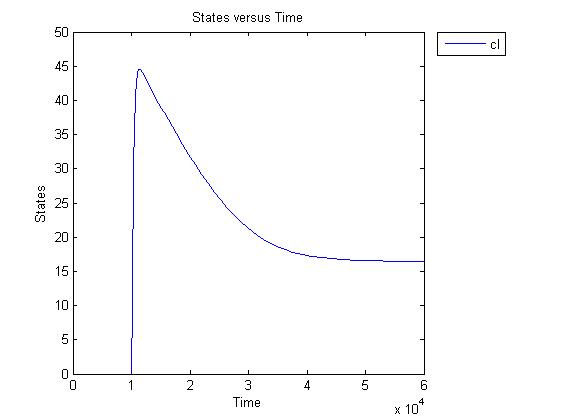
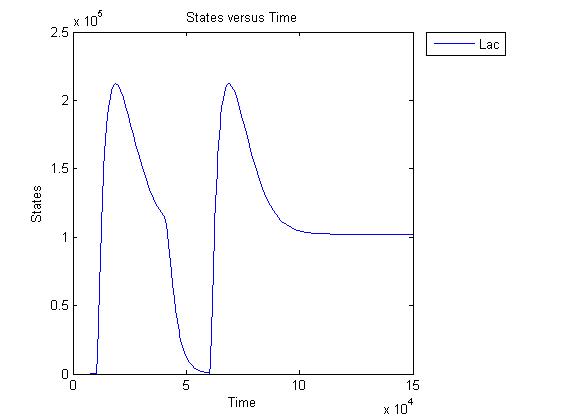
Pulse generator has been reviewed, the parameters have been checked but there are issues... The pulse generator doesn't work (pure physically): when a step signal of T7 is applied cI is produced, which will then repress its transcription. Of course, this will develop to a steady state with a cI level equal to a non zero value. A zero cI value would result in zero repression, thereby restarting the transcription of the cI gene and start producing cI again anyway. Because the cI value remains non zero, so will the lactonase production, and so there will always be a constant amount of lactonase present. Question is: how much will this concentration affect the system considering the case of no light present? No light present means HSL production, which should bind to LuxR, but instead will react with lactonase to an hydroxy-acid. If the concentration of lactonase is considerable, and remains somewhat steady, cell death would never occur. Interesting link to a pulse generator system that seems to work: Pulse Generator Keasling Lab
The lactonase amount does receed to 0 when the filter has 0 output, when the inverter gives high input, but this could cause delay to cell death, due to the slow degradation of lactonase.
The memory system has been reviewed as well. The old system seemed unsuccesful, a new system has been found proven to work. There are however not enough repressors available to implement this system without having interference with other parts. ...
Wiki has been updated a bit more. SBMLSqueezer was used to produce pdf files with the celldesigner reactions, species and parameters. Biobricks pictures were added and some other stuff.
Remarks
iGEM gave us plasmids in cells. Which cells?
Quote of the day
GAS!
 "
"