Team:University of Lethbridge/Notebook/Project2July
From 2008.igem.org
(New page: ===July 28, 2008=== ====Nathan, Roxanne==== Objective: Amplify Riboswitch sequence from pTopp plasmid using PCR Reaction PCR reaction setup: -Enzyme = Phusion -Template = pTopp plasmid...) |
Munima.alam (Talk | contribs) m |
||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Team:University_of_Lethbridge/Notebook|Back to The University of Lethbridge Main Notebook]] | ||
| + | |||
===July 28, 2008=== | ===July 28, 2008=== | ||
| - | ====Nathan, Roxanne==== | + | ====Nathan Puhl, Roxanne==== |
Objective: Amplify Riboswitch sequence from pTopp plasmid using PCR Reaction | Objective: Amplify Riboswitch sequence from pTopp plasmid using PCR Reaction | ||
| Line 14: | Line 16: | ||
PCR cycle conditions (programmed into Brent's PCR machine under "iGEM": | PCR cycle conditions (programmed into Brent's PCR machine under "iGEM": | ||
| - | 1. Initial denaturation @ 98 C for 30 sec (1 cycle) | + | 1. Initial denaturation @ 98 C for 30 sec (1 cycle) |
| - | 2a. Denaturation @ 98 C for 30 sec (35 cycles for step #2) | + | |
| - | 2b. Annealing via gradient for 15 sec | + | 2a. Denaturation @ 98 C for 30 sec (35 cycles for step #2) |
| - | + | ||
| - | + | 2b. Annealing via gradient for 15 sec (45.0 C to 53.0 C) | |
| - | 2c. Extension @ 72 C for 30 sec | + | |
| - | 3. Final extension @ 72 C for 7 mins (1 cycle) | + | 2c. Extension @ 72 C for 30 sec |
| - | 4. Hold at 4 C | + | |
| + | 3. Final extension @ 72 C for 7 mins (1 cycle) | ||
| + | |||
| + | 4. Hold at 4 C | ||
| + | |||
===July 29, 2008=== | ===July 29, 2008=== | ||
====Roxanne, Selina==== | ====Roxanne, Selina==== | ||
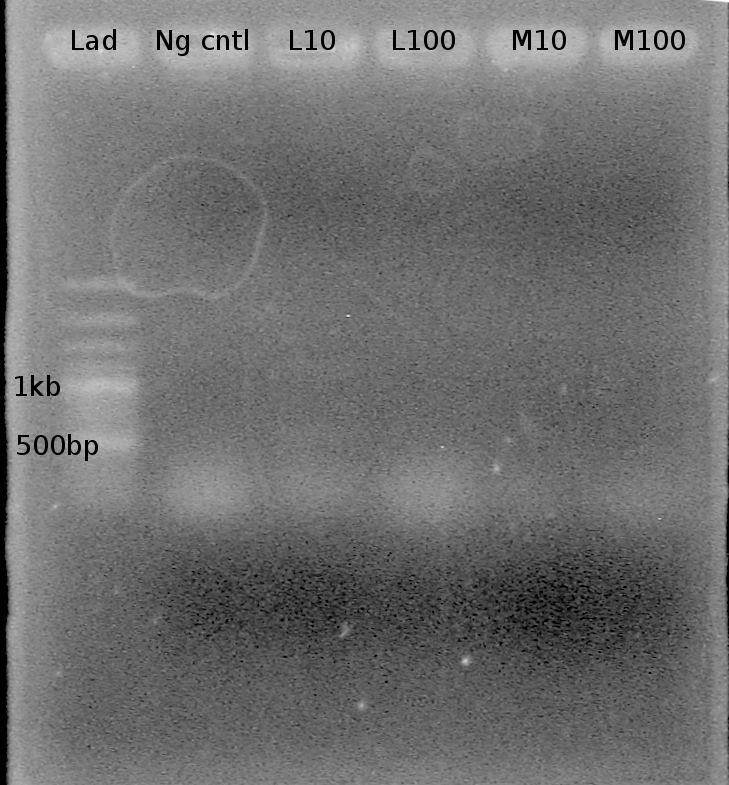
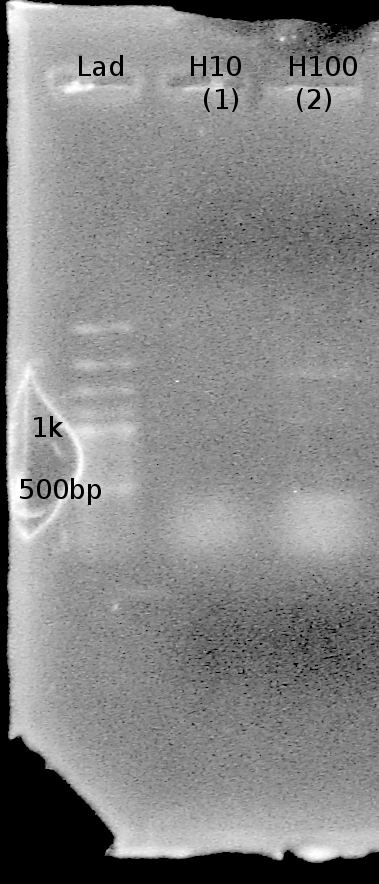
| - | Objective: Visualize whether any Ribswitch PCR reactions (July 28) were successful. | + | Objective: Visualize whether any Ribswitch PCR reactions (July 28) were successful. Expect to see a band around 75 bp. |
| - | Ran 1% TAE agarose gel at 100 V for 30 mins Loaded 5 uL sample, 1 uL ladder | + | Ran 1% TAE agarose gel at 100 V for 30 mins |
| + | |||
| + | Loaded 5 uL sample, 1 uL ladder | ||
Samples: | Samples: | ||
| - | Gel #1: | + | Gel #1: |
| + | - Lane 1: 100 bp ladder | ||
| + | - Lane 2: Neg cntl | ||
| + | - Lane 3: 1:10 template dilution, annealing temp (45.0 C) | ||
| + | - Lane 4: 1:100 template dilution, annealing temp (45.0 C) | ||
| + | - Lane 5: 1:10 template dilution, annealing temp (49.0 C) | ||
| + | - Lane 6: 1:100 template dilution, anneling tenp (49.0 C) | ||
| + | |||
| + | Gel #2: | ||
| + | - Lane 1: 100 bp ladder | ||
| + | - Lane 2: 1:10 template dilution, annealing temp (53.0 C) | ||
| + | - Lane 3: 1:100 template dilution, annealing temp (53.0 C) | ||
| + | |||
| + | Results? Bands present in all lanes! (all around 100 bp) However, unsure if the bands are primer bands or Riboswitch sequence DNA bands. Nathan to run a 3% Agarose gel tonight to try and get better resolution. | ||
| - | + | note: Double bands present in this gel as well. Two bands at ~1kb and 1.5 kb are present in lanes L10 and H100. | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | [[Image:08Jul28_RSPCR.jpg|250 px]] | |
| + | [[Image:08Jul28_RSPCR2.jpg|125 px]] | ||
Latest revision as of 02:34, 30 October 2008
Back to The University of Lethbridge Main Notebook
Contents |
July 28, 2008
Nathan Puhl, Roxanne
Objective: Amplify Riboswitch sequence from pTopp plasmid using PCR Reaction
PCR reaction setup:
-Enzyme = Phusion -Template = pTopp plasmid, dilution series of 1:10, 1:100 and (-) control -Buffer = HF Phusion buffer -Primers = IDT, designed & shipped July 16/08 -Total # of reactions = 20
PCR cycle conditions (programmed into Brent's PCR machine under "iGEM":
1. Initial denaturation @ 98 C for 30 sec (1 cycle)
2a. Denaturation @ 98 C for 30 sec (35 cycles for step #2)
2b. Annealing via gradient for 15 sec (45.0 C to 53.0 C)
2c. Extension @ 72 C for 30 sec
3. Final extension @ 72 C for 7 mins (1 cycle)
4. Hold at 4 C
July 29, 2008
Roxanne, Selina
Objective: Visualize whether any Ribswitch PCR reactions (July 28) were successful. Expect to see a band around 75 bp.
Ran 1% TAE agarose gel at 100 V for 30 mins
Loaded 5 uL sample, 1 uL ladder
Samples:
Gel #1: - Lane 1: 100 bp ladder - Lane 2: Neg cntl - Lane 3: 1:10 template dilution, annealing temp (45.0 C) - Lane 4: 1:100 template dilution, annealing temp (45.0 C) - Lane 5: 1:10 template dilution, annealing temp (49.0 C) - Lane 6: 1:100 template dilution, anneling tenp (49.0 C)
Gel #2: - Lane 1: 100 bp ladder - Lane 2: 1:10 template dilution, annealing temp (53.0 C) - Lane 3: 1:100 template dilution, annealing temp (53.0 C)
Results? Bands present in all lanes! (all around 100 bp) However, unsure if the bands are primer bands or Riboswitch sequence DNA bands. Nathan to run a 3% Agarose gel tonight to try and get better resolution.
note: Double bands present in this gel as well. Two bands at ~1kb and 1.5 kb are present in lanes L10 and H100.
 "
"