Team:Caltech/Protocols/PABA HPLC assay
From 2008.igem.org
(Difference between revisions)
(→Sources) |
|||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Caltech_iGEM_08| | |
| - | + | Content= | |
| - | + | ||
| - | {{Caltech_iGEM_08 | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <div style="font-size:18pt;"> | ||
| + | <font face="verdana" style="color:#CC3300">para-Aminobenzoic Acid (pABA) HPLC Assay Protocol</font></div> | ||
| + | <br> | ||
| + | __TOC__ | ||
==Purpose== | ==Purpose== | ||
| - | To test for para-aminobenzoic acid levels in 'E. coli' overexpressing the pABA synthesis genes 'pabA' and 'pabB'. | + | To test for para-aminobenzoic acid levels in ''E. coli'' overexpressing the pABA synthesis genes ''pabA'' and ''pabB''. |
==Materials== | ==Materials== | ||
| Line 18: | Line 13: | ||
* Buffer B: 0.1% formic acid in MeOH | * Buffer B: 0.1% formic acid in MeOH | ||
* A high performance liquid chromatography machine. | * A high performance liquid chromatography machine. | ||
| - | * Column | + | * HPLC Column: Eclipse XDB-C18 column, 5um particles, 4.6x150mm column, with attached guard column (all from Agilent) |
==Procedure== | ==Procedure== | ||
===Sample Prep=== | ===Sample Prep=== | ||
| - | + | # Centrifuge the cell culture max speed, 10 minutes. | |
| - | + | # Separate pellet from supernatant. | |
| - | + | # Resuspend the pellet in 1mL 0.1M Tris-HCl buffer | |
| - | + | # Sonicate the resuspended pellet for 1 minute, alternating between 0 and 12Hz. | |
| - | + | # Filter sterilize the cell lysate and the supernatant. | |
| - | + | # Load 500uL of each into the HPLC. | |
===HPLC Method=== | ===HPLC Method=== | ||
| - | + | # Injection volume was 20uL, column temperature was kept at 40°C. | |
| - | + | # Flush the lines individually with Buffer A and B for two minutes each, flush the column with 92% A, 8% B for 10 minutes. | |
| - | + | # From the source: "The starting eluent was 92% A mixed with 8% B; the proportion of B was increased linearly to 50% in 7 min, then to 100% in 3 min. The mobile phase was then immediately adjusted to its initial composition and held for 4 min in order to re-equilibrate the column." | |
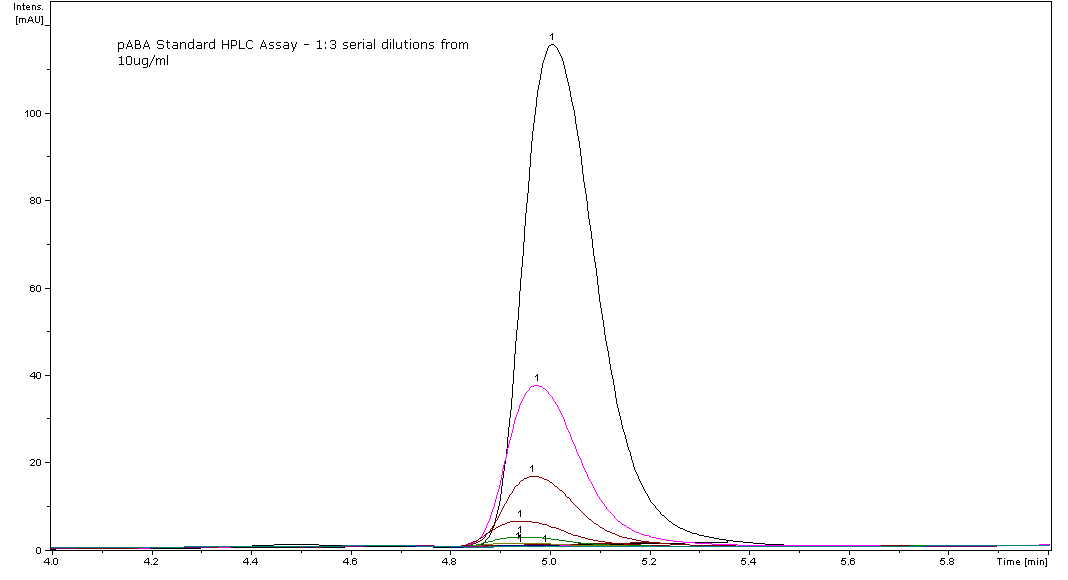
| - | + | # We found the retention time of pABA to be 4.9-5.0 minutes, however the literature states the retention time as 6.0 minutes. | |
| + | |||
| + | [[Image:PABA Standard HPLC assay - 08-07-08.PNG|thumb|center|pABA eluting at 4.9-5.0 minutes for the standard curve.]] | ||
===Standard Curve=== | ===Standard Curve=== | ||
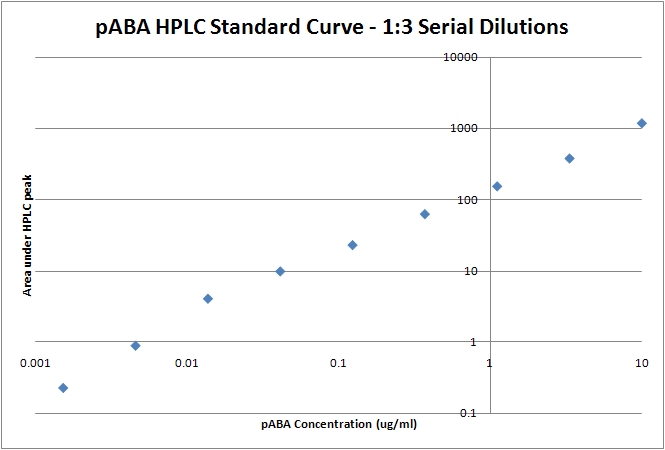
| - | 1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol. | + | 1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol to generate a linear standard (see below). |
| - | ==Sources== | + | [[Image:08-07-08_pABA_Standard_Curve_plot.jpg | thumb|center |pABA HPLC standard curve with 1:3 dilutions starting from 10ug/ml pABA. The integral of the peak is plotted against the known pABA amount.]] |
| + | |||
| + | ===Sources=== | ||
Guo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. ''Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry.'' '''Rapid Communications in Mass Spectrometry''': Volume 19, Issue 8 , Pages 963 - 969, 2005. | Guo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. ''Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry.'' '''Rapid Communications in Mass Spectrometry''': Volume 19, Issue 8 , Pages 963 - 969, 2005. | ||
| + | |||
| + | ===Contacts=== | ||
| + | [[User:vhsiao|Victoria Hsiao]] | ||
| + | }} | ||
Latest revision as of 20:55, 15 October 2008
|
People
|
para-Aminobenzoic Acid (pABA) HPLC Assay Protocol
PurposeTo test for para-aminobenzoic acid levels in E. coli overexpressing the pABA synthesis genes pabA and pabB. Materials
ProcedureSample Prep
HPLC Method
Standard Curve1. Run 1:3 dilutions starting from 10ug/ml pABA in wild type bacterial lysate using the same protocol to generate a linear standard (see below). SourcesGuo-Fang Zhang, Kjell A. Mortier, Sergei Storozhenko, Jet Van De Steene, Dominique Van Der Straeten, Willy E. Lambert. Free and total para-aminobenzoic acid analysis in plants with high-performance liquid chromatography/tandem mass spectrometry. Rapid Communications in Mass Spectrometry: Volume 19, Issue 8 , Pages 963 - 969, 2005. Contacts |
 "
"