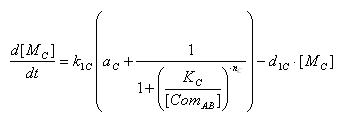Team:NTU-Singapore/Parts/Failed Parameter Estimation
From 2008.igem.org
Lalala8585 (Talk | contribs) (New page: <html><link rel="stylesheet" href="http://edventure.ntu.edu.sg/bbcswebdav/users/z060022/ntu_igem.css" type="text/css"></html> <div id="header">{{User:Greenbear/sandbox/header}}</div> <di...) |
Lalala8585 (Talk | contribs) |
||
| (6 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="header">{{User:Greenbear/sandbox/header}}</div> | <div id="header">{{User:Greenbear/sandbox/header}}</div> | ||
| - | <div id="maincontent" style="margin-top: | + | <div id="maincontent" style="margin-top:100px;"> |
| + | <html> | ||
| + | <div id="arrow"> | ||
| + | <a href="https://2008.igem.org/Team:NTU-Singapore/Parts/Parameter_Estimation_using_Characterization_Results#Parameter_Estimation"> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/6/65/Back_to_corr_copy3.png" | ||
| + | alt="Back to Parameter Estimation & Correlations" | ||
| + | title="Back to Parameter Estimation & Correlations"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </html> | ||
=Introduction= | =Introduction= | ||
'''Transcription of GFP gene mRNA'''<br> | '''Transcription of GFP gene mRNA'''<br> | ||
[[Image:E71.JPG |300px|E71]]<br> | [[Image:E71.JPG |300px|E71]]<br> | ||
| - | [MC]: | + | {|border="0"| |
| - | k1C: | + | |[MC]: |
| - | a : constitutive portion , 0<a<1 | + | |E7 mRNA Concentration |
| - | KC: | + | |- |
| - | nC: | + | |k1C: |
| - | d1C: degradation constant for mRNA | + | |kinetic constant of transcription |
| + | |- | ||
| + | |a : | ||
| + | |constitutive portion , 0<a<1 | ||
| + | |- | ||
| + | |KC: | ||
| + | |Hill constant | ||
| + | |- | ||
| + | |nC: | ||
| + | |Hill coefficient | ||
| + | |- | ||
| + | |d1C: | ||
| + | |degradation constant for mRNA | ||
| + | |} | ||
'''Translation of GFP Protein'''<br> | '''Translation of GFP Protein'''<br> | ||
[[Image:E72.JPG |200px|E72]]<br> | [[Image:E72.JPG |200px|E72]]<br> | ||
| - | [PC] : Protein concentration | + | {|border="0"| |
| - | k2C : kinetic constant of translation | + | |[PC] : |
| - | d2C : degradation constant for Protein | + | |Protein concentration |
| - | + | |- | |
| + | |k2C : | ||
| + | |kinetic constant of translation | ||
| + | |- | ||
| + | |d2C : | ||
| + | |degradation constant for Protein | ||
| + | |} | ||
Above shown are the parameters of interest in this estiamtion exercise<br> | Above shown are the parameters of interest in this estiamtion exercise<br> | ||
| Line 29: | Line 57: | ||
=Parameter Estimation Results= | =Parameter Estimation Results= | ||
| - | [[Image:AC.jpg|800px|aC]]<br> | + | [[Image:AC.jpg|800px|thumb|center|Parameter Estimation Results of aC]]<br> |
| - | [[Image:D1C.jpg|800px|d1C]]<br> | + | [[Image:D1C.jpg|800px|thumb|center|Parameter Estimation Results of d1C]]<br> |
| - | [[Image:D2C.jpg|800px|d2C]]<br> | + | [[Image:D2C.jpg|800px|thumb|center|Parameter Estimation Results of d2C]]<br> |
| - | [[Image:K1C.jpg|800px|k1C]]<br> | + | [[Image:K1C.jpg|800px|thumb|center|Parameter Estimation Results of k1C]]<br> |
| - | [[Image:K2C.jpg|800px|k2C]]<br> | + | [[Image:K2C.jpg|800px|thumb|center|Parameter Estimation Results of k2C]]<br> |
| - | [[Image:KC.jpg|800px|KC]]<br> | + | [[Image:KC.jpg|800px|thumb|center|Parameter Estimation Results of KC]]<br> |
| - | [[Image:NC.jpg|800px|nC]]<br> | + | [[Image:NC.jpg|800px|thumb|center|Parameter Estimation Results of nC]]<br> |
As seen the results are not consistent at all. It was assumed that the fluoresence observed would be directly proportional to the amount of proteins formed. The above results show that was not the case. | As seen the results are not consistent at all. It was assumed that the fluoresence observed would be directly proportional to the amount of proteins formed. The above results show that was not the case. | ||
| + | |||
| + | <html> | ||
| + | <div id="arrow"> | ||
| + | <a href="https://2008.igem.org/Team:NTU-Singapore/Parts/Parameter_Estimation_using_Characterization_Results#Parameter_Estimation"> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/2/2d/Back_to_param_charac.png" | ||
| + | alt="Back to Parameter Estimation" | ||
| + | title="Back to Parameter Estimation"> | ||
| + | </a> | ||
| + | </div> | ||
| + | </html> | ||
| + | <br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| + | |||
| + | var tags_before_clock = "<b>It is now " | ||
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.write('<layer id="clock"></layer><br>'); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | document.write('<span id="clock"></span>'); | ||
| + | } | ||
| + | |||
| + | DaysofWeek = new Array() | ||
| + | DaysofWeek[0]="Sunday" | ||
| + | DaysofWeek[1]="Monday" | ||
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| + | |||
| + | Months = new Array() | ||
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | setInterval("upclock()",1000); | ||
| + | //--> | ||
| + | </script> | ||
| + | </html> | ||
Latest revision as of 07:22, 27 October 2008
|
Introduction
Transcription of GFP gene mRNA
| [MC]: | E7 mRNA Concentration |
| k1C: | kinetic constant of transcription |
| a : | constitutive portion , 0<a<1 |
| KC: | Hill constant |
| nC: | Hill coefficient |
| d1C: | degradation constant for mRNA |
| [PC] : | Protein concentration |
| k2C : | kinetic constant of translation |
| d2C : | degradation constant for Protein |
Above shown are the parameters of interest in this estiamtion exercise
Parameter estimation of the constants in the transcription and translation of the GFP protein was performed using the RFU results obtained. The estimations were done using [http://www.mathworks.com/access/helpdesk/help/toolbox/slestim/index.html?/access/helpdesk/help/toolbox/slestim/rn/bqnp6nw-1.html&http://www.google.com/search?q=simulink+parameter+estimation&rls=com.microsoft:*&ie=UTF-8&oe=UTF-8&startIndex=&startPage=1 Simulink Parameter Estimation]. The approach was overly simplistic but done to get a feel of how the parameters would respond.
Parameter Estimation Results
As seen the results are not consistent at all. It was assumed that the fluoresence observed would be directly proportional to the amount of proteins formed. The above results show that was not the case.
 "
"