Imperial College/1 September 2008
From 2008.igem.org
(Difference between revisions)
m (→1st September 2008) |
|||
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{| cellpadding="10" border="0" | {| cellpadding="10" border="0" | ||
|- valign="top" | |- valign="top" | ||
| - | |||
|{{#calendar: title=Imperial_College |year=2008 | month=08}} | |{{#calendar: title=Imperial_College |year=2008 | month=08}} | ||
|{{#calendar: title=Imperial_College |year=2008 | month=09}} | |{{#calendar: title=Imperial_College |year=2008 | month=09}} | ||
| - | | rowspan="2" bgcolor=# | + | |{{#calendar: title=Imperial_College |year=2008 | month=10}} |
| + | | rowspan="2" bgcolor=#ffffff width="100%" | | ||
|} | |} | ||
| + | =1 September 2008= | ||
==Wet Lab== | ==Wet Lab== | ||
| - | |||
| - | |||
| - | |||
===Cloning=== | ===Cloning=== | ||
| - | + | *Trial PCRs to obtain the optimal conditions of our primers for vector template DNA cloning were set up and run according to the standard [http://openwetware.org/index.php?title=IGEM:IMPERIAL/2008/Prototype/Wetlab/PCR protocol] (results below) | |
* First GeneArt sequences arrived! Constructs 7, 8 9 and 11 (SacB-EAK 16, LipA-EAK16, P43-gsiB and Pveg-gsiB) transformed into XL1-Blue ''E.coli'' | * First GeneArt sequences arrived! Constructs 7, 8 9 and 11 (SacB-EAK 16, LipA-EAK16, P43-gsiB and Pveg-gsiB) transformed into XL1-Blue ''E.coli'' | ||
| Line 22: | Line 20: | ||
Results from the growth curve trial are shown below: | Results from the growth curve trial are shown below: | ||
{| align=center | {| align=center | ||
| - | ! Time (hours) || Actual Time || 1ml OD< | + | ! Time (hours) || Actual Time || 1ml OD<sub>600</sub> || 10ml OD<sub>600</sub> || Plated? || Dilutions to Use |
|- | |- | ||
| - | | 0 || 0940 || 0.057 || 0.006 || N | + | | 0 || 0940 || 0.057 || 0.006 || N || |
|- | |- | ||
| - | | 0.5 || 1020 || 0.055 || 0.006 || N | + | | 0.5 || 1020 || 0.055 || 0.006 || N || |
|- | |- | ||
| - | | 1 || 1050 || 0.054 || 0.003 || Y | + | | 1 || 1050 || 0.054 || 0.003 || Y || |
|- | |- | ||
| - | | 2 || 1145 || 0.062 || 0.008 || Y | + | | 2 || 1145 || 0.062 || 0.008 || Y || x10<sup>4-6</sup> |
|- | |- | ||
| - | | 4 || 1340 || 0.531 || 0.066 || N | + | | 4 || 1340 || 0.531 || 0.066 || N || |
|- | |- | ||
| - | | 8 || 1745 || 1.390 || 0.232 || Y | + | | 8 || 1745 || 1.390 || 0.232 || Y || x10<sup>6-8</sup> |
|- | |- | ||
| - | | 24 || 1000 || 1.762 || 0.305 || Y | + | | 24 || 1000 || 1.762 || 0.305 || Y || x10<sup>5-9</sup> |
|} | |} | ||
| + | |||
| + | ==Debriefing== | ||
| + | Today we had an extensive debreifing session, the notes below summaries the key points: | ||
| + | *Erika and Prudence - Working on the ODE's of the genetic circuits. Studying the testing parts, looking for the individual models that can help to contribute the final model of the genetic circuit. | ||
| + | *Clinton and Yanis - Carried out statistical evaluation of the two tracking programs. The sum of there findings is that will use the manual software. However, using this software the tumbling events were manually put in, this was based on the assumption that any curved deviating swimming pattern was the tumbling events. This assumption clearly needs to be validated, as a result now a search into the swimming patterns of ''B.subtilis'' will be carried out. This includes liertature and contacting relevant experts to get an approximation of the swimming time. In addition the group has started to think about the data that will be collected, this includes distribution curves for velocity, tumbling times, running times. For the characterisation of wild type swimming the group feels that by the end of next week they will have there final distribution curves for these. In addition the type of characterisation of EpsE effect on motility as discussed. | ||
| + | *Dry lab group - By the end of next week half the dry lab team is leaving, this means there is an urgency for planning of the next two weeks in order to achieve what is required. The dry lab group will make clear aims and targets that need to be reached which will be discussed at the debreif at 5. | ||
| + | *Motility - Plan to conduct data analysis on the motility of wild type B.Subtilis. Distributions of motility parameters will be determined and we may assume that EpsE does not change motility distribution but simply its parameters. | ||
| + | *Wet lab group - 4 of the consititutive promoter and RBS combinations from geneart arrived. In addition to getting these constructs biobricked the group also have to work on PCR to get relevant ''B.subtilis'' parts from the genome and vectors, start second trial on the growth curve and begin constructing parts. A more extensive weekly plan will be uploaded tomorrow. | ||
| + | *Wiki- An internal deadline has been set, this is to upload all the information required on the new blank wiki by saturday, then in the following week the whole group will push towards getting everything checked and apllying a format so that at week 10 we have a 1st draft of our wiki. | ||
| + | |||
| + | ==Results== | ||
| + | |||
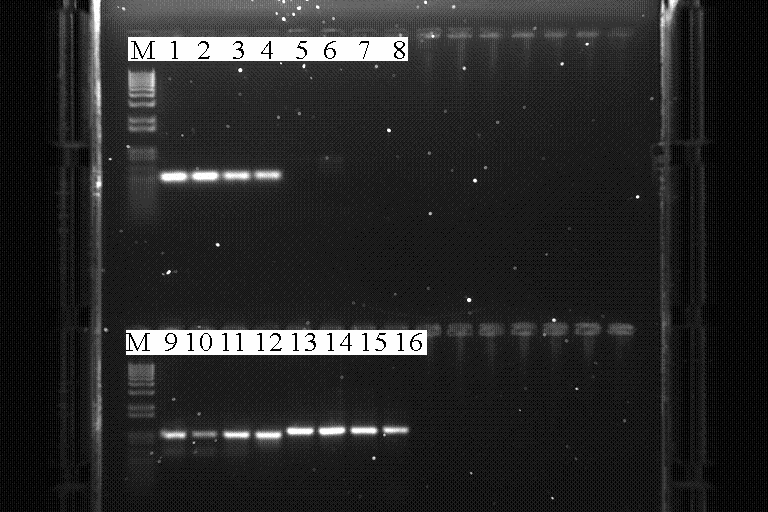
| + | [[Image:1-08PCR.PNG|thumb|600px|center|A 1% Agarose gel showing the results of various PCR reactions, M = Marker]] | ||
| + | |||
| + | *Lane 1- First 10 cycles at 54<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using AmyE 5' integration sequence primers, (effectively a positive control) | ||
| + | *Lane 2- First 10 cycles at 52<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using AmyE 5' integration sequence primers, | ||
| + | *Lane 3- First 10 cycles at 50<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using AmyE 5' integration sequence primers, | ||
| + | *Lane 4- First 10 cycles at 48<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using AmyE 5' integration sequence primers, | ||
| + | <br> | ||
| + | *Lane 5- First 10 cycles at 54<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using Aad9 primers, | ||
| + | *Lane 6- First 10 cycles at 52<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using Aad9 primers, | ||
| + | *Lane 7- First 10 cycles at 50<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using Aad9 primers, | ||
| + | *Lane 8- First 10 cycles at 48<sup>o</sup>C and then 20 cycles at 60<sup>o</sup>C using Aad9 primers, | ||
| + | <br> | ||
| + | *Lane 9- First 10 cycles at 58<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using AmyE 3' integration sequence primers, | ||
| + | *Lane 10- First 10 cycles at 56<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using AmyE 3' integration sequence primers, | ||
| + | *Lane 11- First 10 cycles at 54<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using AmyE 3' integration sequence primers, | ||
| + | *Lane 12- First 10 cycles at 52<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using AmyE 3' integration sequence primers, | ||
| + | <br> | ||
| + | *Lane 13- First 10 cycles at 58<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using LacI gene primers, | ||
| + | *Lane 14- First 10 cycles at 56<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using LacI gene primers, | ||
| + | *Lane 15- First 10 cycles at 54<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using LacI gene primers, | ||
| + | *Lane 16- First 10 cycles at 52<sup>o</sup>C and then 20 cycles at 65<sup>o</sup>C using LacI gene primers, | ||
| + | <br> | ||
| + | *Optimal conditions have been determined for the primers for both AmyE integration sequences and LacI adn so can be used in Pfu cloning reactions tomorrow. Further testing will have to be undertaken with Taq for Aad9 | ||
| + | <br> | ||
| + | {{Imperial/EndPage|Notebook|Notebook}} | ||
Latest revision as of 20:43, 28 October 2008
1 September 2008Wet LabCloning
Growth CurveThe illuminator's software is not sensitive enough to detect colonies accurately to count them. We have decided to count colonies manually instead. Results from the growth curve trial are shown below:
DebriefingToday we had an extensive debreifing session, the notes below summaries the key points:
Results
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"