Team:Guelph/Modeling
From 2008.igem.org
m |
|||
| (2 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
!style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Parts|<font color="#cd0000">Parts</font>]] | !style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Parts|<font color="#cd0000">Parts</font>]] | ||
!style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Notebook|<font color="#cd0000">Notebook</font>]] | !style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Notebook|<font color="#cd0000">Notebook</font>]] | ||
| - | !style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/ | + | !style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Results|<font color="#cd0000">Results</font>]] |
!style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Links|<font color="#cd0000">Links</font>]] | !style="font-size:140%;text-align:center; background-color:#ffff00; border-width:0px; padding:3px;"|[[Team:Guelph/Links|<font color="#cd0000">Links</font>]] | ||
|} | |} | ||
| Line 27: | Line 27: | ||
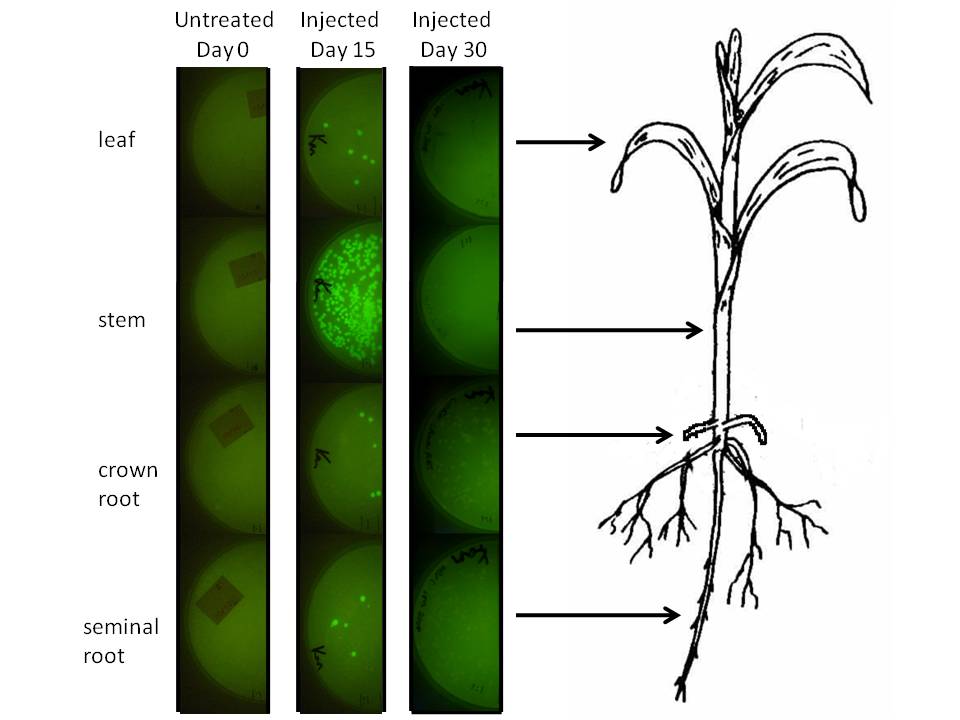
In corn, we are doing this using a corn endophyte called Klebsiella pneumonii. Plants were either injected with 5 ul of bacterial suspension or dipped in it at an early stage. Two weeks later (a month for the dipped plants) 500 mg of tissue were harvested, ground in sterilized mortars, resuspended in 500 ul of sodium phosphate buffer, and 50 ul of the dilution was spread on half of a kanamycin LB plate. The results are depicted graphically below. | In corn, we are doing this using a corn endophyte called Klebsiella pneumonii. Plants were either injected with 5 ul of bacterial suspension or dipped in it at an early stage. Two weeks later (a month for the dipped plants) 500 mg of tissue were harvested, ground in sterilized mortars, resuspended in 500 ul of sodium phosphate buffer, and 50 ul of the dilution was spread on half of a kanamycin LB plate. The results are depicted graphically below. | ||
| - | [[Image: | + | [[Image:KP342 GFP assay in corn.jpg|700px]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 05:32, 27 September 2008
| Home | The Team | The Project | Parts | Notebook | Results | Links |
|---|
Making projections based on measurements on endosymbiont and endophyte numbers within a functioning biological system using GFP
We plan to do wet lab modelling. By tagging our microbes with simple GFP constructs and counting colony forming units per set sample fresh weight, we hope to be able to observe microbial survival with our construct and predict how much beta carotene or RNAi might be produced and released.
In corn, we are doing this using a corn endophyte called Klebsiella pneumonii. Plants were either injected with 5 ul of bacterial suspension or dipped in it at an early stage. Two weeks later (a month for the dipped plants) 500 mg of tissue were harvested, ground in sterilized mortars, resuspended in 500 ul of sodium phosphate buffer, and 50 ul of the dilution was spread on half of a kanamycin LB plate. The results are depicted graphically below.
 "
"