Imperial College/9 September 2008
From 2008.igem.org
m (New page: =8 September 2008= =='''Wet Lab'''== *Taq PCR trials of genomic sequences set up and results analysed on gel *Ligations of all available sequences into biobricks set up [[Image:9-08PCR...) |
|||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | = | + | {{Imperial/StartPage}}__NOTOC__ |
| + | {| cellpadding="10" border="0" | ||
| + | |- valign="top" | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=08}} | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=09}} | ||
| + | |{{#calendar: title=Imperial_College |year=2008 | month=10}} | ||
| + | | rowspan="2" bgcolor=#ffffff width="100%" | | ||
| + | |} | ||
| + | =9 September 2008= | ||
| Line 9: | Line 17: | ||
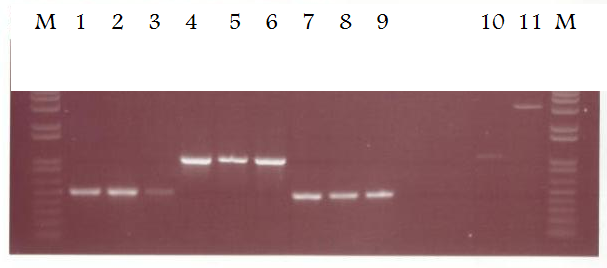
[[Image:9-08PCR.PNG|thumb|400px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer. M = Marker]] | [[Image:9-08PCR.PNG|thumb|400px|center|A 1% Agarose gel showing the results of various PCR reactions. Each lane is loaded with 5ul of PCR reaction and 1ul of 6x sample buffer. M = Marker]] | ||
| - | Lane 1 - EpsE 5' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 1 - EpsE 5' IS primers, chromosome template, 52<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
| - | Lane 2 - EpsE 5' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 2 - EpsE 5' IS primers, chromosome template, 50<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
| - | Lane 3 - EpsE 5' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 3 - EpsE 5' IS primers, chromosome template, 48<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
| - | Lane 4 - XylR primers, chromosome template, <sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles | + | Lane 4 - XylR primers, chromosome template, 50<sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles |
| - | Lane 5 - XylR primers, chromosome template, <sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles | + | Lane 5 - XylR primers, chromosome template, 48<sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles |
| - | Lane 6 - XylR primers, chromosome template, <sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles | + | Lane 6 - XylR primers, chromosome template, 46<sup>o</sup>C for 10 cycles then 60<sup>o</sup>C for 20 cycles |
| - | Lane 7 - EpsE 3' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 7 - EpsE 3' IS primers, chromosome template, 54<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
| - | Lane 8 - EpsE 3' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 8 - EpsE 3' IS primers, chromosome template, 52<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
| - | Lane 9 - EpsE 3' IS primers, chromosome template, <sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles | + | Lane 9 - EpsE 3' IS primers, chromosome template, 50<sup>o</sup>C for 10 cycles then 64<sup>o</sup>C for 20 cycles |
Lane 10 - LacI insert (cut with ''Xba''I and ''Spe''I) | Lane 10 - LacI insert (cut with ''Xba''I and ''Spe''I) | ||
| Line 31: | Line 39: | ||
Lane 11 - pSB1AK3 vector (cut with ''Xba''I and ''Spe''I) | Lane 11 - pSB1AK3 vector (cut with ''Xba''I and ''Spe''I) | ||
| - | Conditions and timings were kept the same as the standard Taq protocol. Pfu PCR reactions using the optimal condiitons can now be set up to produce clones for use in our constructs. From the LacI and pSB1AK3 digests, it is now possible to decide how much of each is required in our ligation reaction | + | Conditions and timings were kept the same as the standard Taq protocol. Pfu PCR reactions using the optimal condiitons can now be set up to produce clones for use in our constructs. From the LacI and pSB1AK3 digests, it is now possible to decide how much of each is required in our ligation reaction. |
| + | |||
| + | <br> | ||
| + | {{Imperial/EndPage|Notebook|Notebook}} | ||
Latest revision as of 20:45, 28 October 2008
9 September 2008Wet Lab
Lane 1 - EpsE 5' IS primers, chromosome template, 52oC for 10 cycles then 64oC for 20 cycles Lane 2 - EpsE 5' IS primers, chromosome template, 50oC for 10 cycles then 64oC for 20 cycles Lane 3 - EpsE 5' IS primers, chromosome template, 48oC for 10 cycles then 64oC for 20 cycles Lane 4 - XylR primers, chromosome template, 50oC for 10 cycles then 60oC for 20 cycles Lane 5 - XylR primers, chromosome template, 48oC for 10 cycles then 60oC for 20 cycles Lane 6 - XylR primers, chromosome template, 46oC for 10 cycles then 60oC for 20 cycles Lane 7 - EpsE 3' IS primers, chromosome template, 54oC for 10 cycles then 64oC for 20 cycles Lane 8 - EpsE 3' IS primers, chromosome template, 52oC for 10 cycles then 64oC for 20 cycles Lane 9 - EpsE 3' IS primers, chromosome template, 50oC for 10 cycles then 64oC for 20 cycles Lane 10 - LacI insert (cut with XbaI and SpeI) Lane 11 - pSB1AK3 vector (cut with XbaI and SpeI) Conditions and timings were kept the same as the standard Taq protocol. Pfu PCR reactions using the optimal condiitons can now be set up to produce clones for use in our constructs. From the LacI and pSB1AK3 digests, it is now possible to decide how much of each is required in our ligation reaction.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"