Team:NTU-Singapore/Parts
From 2008.igem.org
Lalala8585 (Talk | contribs) (→Characterization of Standard Biobrick via GFP Activity) |
Lalala8585 (Talk | contribs) |
||
| (15 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<div id="body" style="margin-top:200px;width:950px;padding:5px;"> | <div id="body" style="margin-top:200px;width:950px;padding:5px;"> | ||
| - | = | + | ='''Characterization Introduction'''= |
The group proposed to discuss and investigate the working range of the parts and devices of our system | The group proposed to discuss and investigate the working range of the parts and devices of our system | ||
*The proposed variables to investigate are: <br> | *The proposed variables to investigate are: <br> | ||
| Line 11: | Line 11: | ||
i) Concentration of activator<br> | i) Concentration of activator<br> | ||
| - | ii) | + | ii) Temperature <br> |
| - | + | ||
| - | iii) | + | iii) Time <br> |
==Characterization of Parts== | ==Characterization of Parts== | ||
| Line 22: | Line 22: | ||
ii) Iron Sensitive Promoter pFe [http://partsregistry.org/wiki/index.php?title=Part:BBa_I716014 BBa_I716014] | ii) Iron Sensitive Promoter pFe [http://partsregistry.org/wiki/index.php?title=Part:BBa_I716014 BBa_I716014] | ||
| - | iii) [ | + | iii) AI2 senstive Promoter pLsrA [http://partsregistry.org/wiki/index.php?title=Part:BBa_K117002 BBa_K117002] <br> |
The characterization is carried out by ligating the desired promoter with the Green Fluorescence protein: [http://partsregistry.org/Part:BBa_E0240 BBa_E02040] | The characterization is carried out by ligating the desired promoter with the Green Fluorescence protein: [http://partsregistry.org/Part:BBa_E0240 BBa_E02040] | ||
*For the pLac BBa_R0010, it is already available in the form that we desired in the registry as [http://partsregistry.org/wiki/index.php?title=Part:BBa_J04430 BBa_J04430] | *For the pLac BBa_R0010, it is already available in the form that we desired in the registry as [http://partsregistry.org/wiki/index.php?title=Part:BBa_J04430 BBa_J04430] | ||
*For the pLsrA promoter, the Yellow Fluorescence protein was used instead to differentiate between our two different systems. | *For the pLsrA promoter, the Yellow Fluorescence protein was used instead to differentiate between our two different systems. | ||
| - | == | + | |
| + | =='''Experiments'''== | ||
| + | ==='''1) Characterization of existing LacI regulated promoter (pLacI) via GFP Activity'''=== | ||
[[image: Ntu@iGEM_FLx_800_pic.jpg|400px|center|thumb|The FLx800™ Fluorescence Microplate Reader]] | [[image: Ntu@iGEM_FLx_800_pic.jpg|400px|center|thumb|The FLx800™ Fluorescence Microplate Reader]] | ||
The FLx800™ Fluorescence Microplate Reader was available for our use courtesy of Division of Bio-engineering lab. It was purchased from BioTek and for more information about the reader, you can refer to [http://www.biotek.com/products/product_detail.php?pid=117 The FLx800™ Fluorescence Microplate Reader] | The FLx800™ Fluorescence Microplate Reader was available for our use courtesy of Division of Bio-engineering lab. It was purchased from BioTek and for more information about the reader, you can refer to [http://www.biotek.com/products/product_detail.php?pid=117 The FLx800™ Fluorescence Microplate Reader] | ||
| - | The FLx800™ Fluorescence Microplate Reader has a computer software KCJunior that was installed in a computer adjacent to the reader for collection and processing of data from our samples. | + | The FLx800™ Fluorescence Microplate Reader has a computer software KCJunior that was installed in a computer adjacent to the reader for collection and processing of data from our samples. It was used for the characterisation of pLacI GFP. |
| + | |||
| - | |||
| - | |||
The objectives of the characterization experiment are to investigate the effects of concentration of inducer, temperature and time on the production of Green Fluorescence protein (GFP) by the Biobrick promoter part being experimented. | The objectives of the characterization experiment are to investigate the effects of concentration of inducer, temperature and time on the production of Green Fluorescence protein (GFP) by the Biobrick promoter part being experimented. | ||
| - | A stronger fluorescence, as measured using | + | A stronger fluorescence, as measured using FLx800™ Fluorescence Microplate Reader will indicate more GFP proteins being synthesised. This also directly corresponds to a higher promoter activity. |
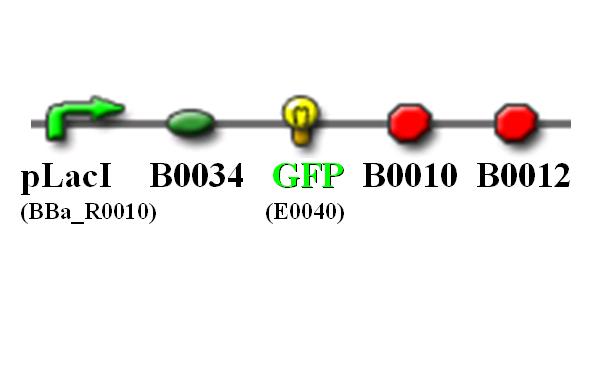
| - | + | Top 10 competent cells are transformed with plasmid carrying the following gene sequence: | |
| - | [[Image: | + | [[Image:NTU@iGEM_PLacI_GFP_pic.JPG|center|thumb|400px|pLacI promoter with GFP reporting gene]] |
The cells are cultured and upon addition of the inducer, the reaction mixtures (with both cells and inducer) are fed into the Fluorescence Microplate Reader. This machine has a inbuilt software, KCJunior, which will translate the results into excellent data suitable for analysis. The samples’ fluorescence was measured for a period of 12 hours, at the temperatures of <br> | The cells are cultured and upon addition of the inducer, the reaction mixtures (with both cells and inducer) are fed into the Fluorescence Microplate Reader. This machine has a inbuilt software, KCJunior, which will translate the results into excellent data suitable for analysis. The samples’ fluorescence was measured for a period of 12 hours, at the temperatures of <br> | ||
i) 25°C <br> | i) 25°C <br> | ||
| Line 46: | Line 47: | ||
and iii) 42°C.<br> | and iii) 42°C.<br> | ||
| - | The readings were than plotted to produce | + | The readings were than plotted to produce 3-dimentional graphs for detail analysis. |
For more information please refer to [https://2008.igem.org/Team:NTU-Singapore/Parts/Characterization_of_LacI-GFP Characterization of pLacI-GFP] | For more information please refer to [https://2008.igem.org/Team:NTU-Singapore/Parts/Characterization_of_LacI-GFP Characterization of pLacI-GFP] | ||
| - | ==Characterization of pLsrA - YFP== | + | ==='''2) Characterization of our New promoter pLsrA-YFP'''=== |
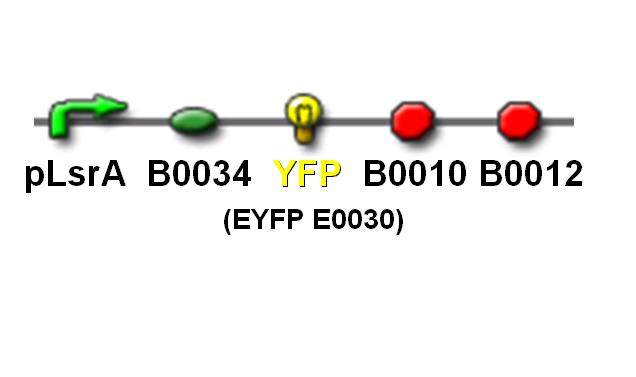
| + | In order to simulate the AND gate detection system, characterization of pLsrA using the Yellow fluorescence protein (YFP) was done. The YFP production is analogous to the Lysis protein production in our AND gate detection system. When the pLsrA promoter is activated by the AI-2 molecules, transcription of the downstream gene, followed by the translation, results in YFP formed. The YFP production is than to be measured using the VICTOR 3 multilabel reader. | ||
| + | [[Image:NTU@iGEM_Victor3-plates-web4web.jpg|center|thumb|400px| VICTOR 3 multilabel reader]]<br> | ||
| + | [[Image: NTU@iGEM_PLsrA_YFP_pic.JPG|center|thumb|400px| pLsrA promoter with YFP reporting gene]]<br> | ||
| + | <br> | ||
| - | For more information please refer to [https://2008.igem.org/Team:NTU-Singapore/Parts/ | + | |
| + | For more information please refer to [https://2008.igem.org/Team:NTU-Singapore/Parts/Characterization_of_pLsrA-YFP Characterization of pLsrA-YFP] | ||
| + | <br><br> | ||
| + | <html> | ||
| + | <script language=Javascript1.2> | ||
| + | <!-- | ||
| + | |||
| + | var tags_before_clock = "<b>It is now " | ||
| + | var tags_middle_clock = "on" | ||
| + | var tags_after_clock = "</b>" | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.write('<layer id="clock"></layer><br>'); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | document.write('<span id="clock"></span>'); | ||
| + | } | ||
| + | |||
| + | DaysofWeek = new Array() | ||
| + | DaysofWeek[0]="Sunday" | ||
| + | DaysofWeek[1]="Monday" | ||
| + | DaysofWeek[2]="Tuesday" | ||
| + | DaysofWeek[3]="Wednesday" | ||
| + | DaysofWeek[4]="Thursday" | ||
| + | DaysofWeek[5]="Friday" | ||
| + | DaysofWeek[6]="Saturday" | ||
| + | |||
| + | Months = new Array() | ||
| + | Months[0]="January" | ||
| + | Months[1]="February" | ||
| + | Months[2]="March" | ||
| + | Months[3]="April" | ||
| + | Months[4]="May" | ||
| + | Months[5]="June" | ||
| + | Months[6]="July" | ||
| + | Months[7]="August" | ||
| + | Months[8]="September" | ||
| + | Months[9]="October" | ||
| + | Months[10]="November" | ||
| + | Months[11]="December" | ||
| + | |||
| + | function upclock(){ | ||
| + | var dte = new Date(); | ||
| + | var hrs = dte.getHours(); | ||
| + | var min = dte.getMinutes(); | ||
| + | var sec = dte.getSeconds(); | ||
| + | var day = DaysofWeek[dte.getDay()] | ||
| + | var date = dte.getDate() | ||
| + | var month = Months[dte.getMonth()] | ||
| + | var year = dte.getFullYear() | ||
| + | |||
| + | var col = ":"; | ||
| + | var spc = " "; | ||
| + | var com = ","; | ||
| + | var apm; | ||
| + | |||
| + | if (date == 1 || date == 21 || date == 31) | ||
| + | {ender = "<sup>st</sup>"} | ||
| + | else | ||
| + | if (date == 2 || date == 22) | ||
| + | {ender = "<sup>nd</sup>"} | ||
| + | else | ||
| + | if (date == 3 || date == 23) | ||
| + | {ender = "<sup>rd</sup>"} | ||
| + | |||
| + | else | ||
| + | {ender = "<sup>th</sup>"} | ||
| + | |||
| + | if (12 < hrs) { | ||
| + | apm="<font size='-1'>pm</font>"; | ||
| + | hrs-=12; | ||
| + | } | ||
| + | |||
| + | else { | ||
| + | apm="<font size='-1'>am</font>"; | ||
| + | } | ||
| + | |||
| + | if (hrs == 0) hrs=12; | ||
| + | if (hrs<=9) hrs="0"+hrs; | ||
| + | if (min<=9) min="0"+min; | ||
| + | if (sec<=9) sec="0"+sec; | ||
| + | |||
| + | if(navigator.appName == "Netscape") { | ||
| + | document.clock.document.write(tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock); | ||
| + | document.clock.document.close(); | ||
| + | } | ||
| + | |||
| + | if (navigator.appVersion.indexOf("MSIE") != -1){ | ||
| + | clock.innerHTML = tags_before_clock+hrs+col+min+col+sec+apm+spc+tags_middle_clock+spc+day+com+spc+date+ender+spc+month+com+spc+year+tags_after_clock; | ||
| + | } | ||
| + | } | ||
| + | |||
| + | setInterval("upclock()",1000); | ||
| + | //--> | ||
| + | </script> | ||
| + | </html> | ||
Latest revision as of 10:15, 27 October 2008
|
Contents |
Characterization Introduction
The group proposed to discuss and investigate the working range of the parts and devices of our system
- The proposed variables to investigate are:
i) Concentration of activator
ii) Temperature
iii) Time
Characterization of Parts
We are interested in characterizing the following parts:
i) Lactose/IPTG sensitive Promoter pLac [http://partsregistry.org/wiki/index.php?title=Part:BBa_R0010 BBa_R0010]
ii) Iron Sensitive Promoter pFe [http://partsregistry.org/wiki/index.php?title=Part:BBa_I716014 BBa_I716014]
iii) AI2 senstive Promoter pLsrA [http://partsregistry.org/wiki/index.php?title=Part:BBa_K117002 BBa_K117002]
The characterization is carried out by ligating the desired promoter with the Green Fluorescence protein: [http://partsregistry.org/Part:BBa_E0240 BBa_E02040]
- For the pLac BBa_R0010, it is already available in the form that we desired in the registry as [http://partsregistry.org/wiki/index.php?title=Part:BBa_J04430 BBa_J04430]
- For the pLsrA promoter, the Yellow Fluorescence protein was used instead to differentiate between our two different systems.
Experiments
1) Characterization of existing LacI regulated promoter (pLacI) via GFP Activity
The FLx800™ Fluorescence Microplate Reader was available for our use courtesy of Division of Bio-engineering lab. It was purchased from BioTek and for more information about the reader, you can refer to [http://www.biotek.com/products/product_detail.php?pid=117 The FLx800™ Fluorescence Microplate Reader]
The FLx800™ Fluorescence Microplate Reader has a computer software KCJunior that was installed in a computer adjacent to the reader for collection and processing of data from our samples. It was used for the characterisation of pLacI GFP.
The objectives of the characterization experiment are to investigate the effects of concentration of inducer, temperature and time on the production of Green Fluorescence protein (GFP) by the Biobrick promoter part being experimented.
A stronger fluorescence, as measured using FLx800™ Fluorescence Microplate Reader will indicate more GFP proteins being synthesised. This also directly corresponds to a higher promoter activity.
Top 10 competent cells are transformed with plasmid carrying the following gene sequence:
The cells are cultured and upon addition of the inducer, the reaction mixtures (with both cells and inducer) are fed into the Fluorescence Microplate Reader. This machine has a inbuilt software, KCJunior, which will translate the results into excellent data suitable for analysis. The samples’ fluorescence was measured for a period of 12 hours, at the temperatures of
i) 25°C
ii) 37°C
and iii) 42°C.
The readings were than plotted to produce 3-dimentional graphs for detail analysis.
For more information please refer to Characterization of pLacI-GFP
2) Characterization of our New promoter pLsrA-YFP
In order to simulate the AND gate detection system, characterization of pLsrA using the Yellow fluorescence protein (YFP) was done. The YFP production is analogous to the Lysis protein production in our AND gate detection system. When the pLsrA promoter is activated by the AI-2 molecules, transcription of the downstream gene, followed by the translation, results in YFP formed. The YFP production is than to be measured using the VICTOR 3 multilabel reader.
For more information please refer to Characterization of pLsrA-YFP
 "
"