Team:Caltech/Project
From 2008.igem.org
Caltechigem (Talk | contribs) |
|||
| (6 intermediate revisions not shown) | |||
| Line 21: | Line 21: | ||
{{!}}-valign="top" | {{!}}-valign="top" | ||
{{!}} | {{!}} | ||
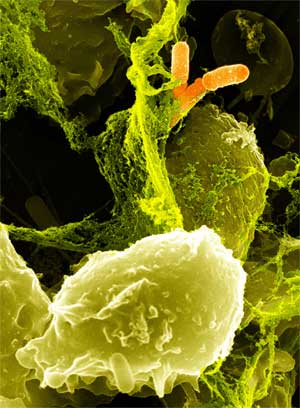
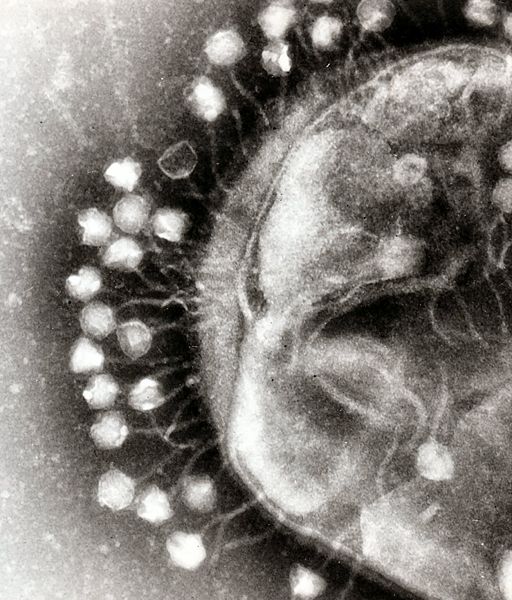
| - | + | Bacterial food poisoning is a prevalent problem around the world. In the year 2006, the United States had more than 325,000 hospitalizations from food borne illnesses. This subproject focuses on the prevention of food poisioning by the release of pathogen-specific bacteriophage. A model system for ‘manufacturing’ phage was built around ''Escherichia coli'' bacteriophage λ lysogens. We began with a strain of ''E. coli'' which does not express the receptor, LamB, necessary for λ infection. Next we expressed the receptor in the cell, allowing infection and lysogeny. Then the receptor was removed, again rendering the cell non-susceptible. Finally, a plasmid was constructed to control the induction of λ by regulating the expression of ''rcsA'' and ''cI''. When fully implemented, this system could be used to treat food poisoning due to pathogenic ''E. coli''. The system can also be modified to target other pathogenic species, such as ''Salmonella'', by replacing λ phage with other temperate bacteriophages and using similar methods to incorporate the non-native phage into ''E. coli''. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{!}}{{navimg|xsize=220|ysize=258|image=Phage.jpg|link=Team:Caltech/Project/Phage_Pathogen_Defense}} | {{!}}{{navimg|xsize=220|ysize=258|image=Phage.jpg|link=Team:Caltech/Project/Phage_Pathogen_Defense}} | ||
{{!}}} | {{!}}} | ||
| Line 53: | Line 49: | ||
As more complicated and interconnected biological circuits are built, there is an increasing need for the simple integration of multiple functions into a single bacterial cell line. However, some of these functions may be incompatible or may kill the cell, such that each cell can only express a single function at any time or must be regenerated if the functionality is fatal. We aim to combine multiple mutually exclusive and potentially fatal functions into a single bacterial cell line that, as a population, exhibits the entire set of functions. | As more complicated and interconnected biological circuits are built, there is an increasing need for the simple integration of multiple functions into a single bacterial cell line. However, some of these functions may be incompatible or may kill the cell, such that each cell can only express a single function at any time or must be regenerated if the functionality is fatal. We aim to combine multiple mutually exclusive and potentially fatal functions into a single bacterial cell line that, as a population, exhibits the entire set of functions. | ||
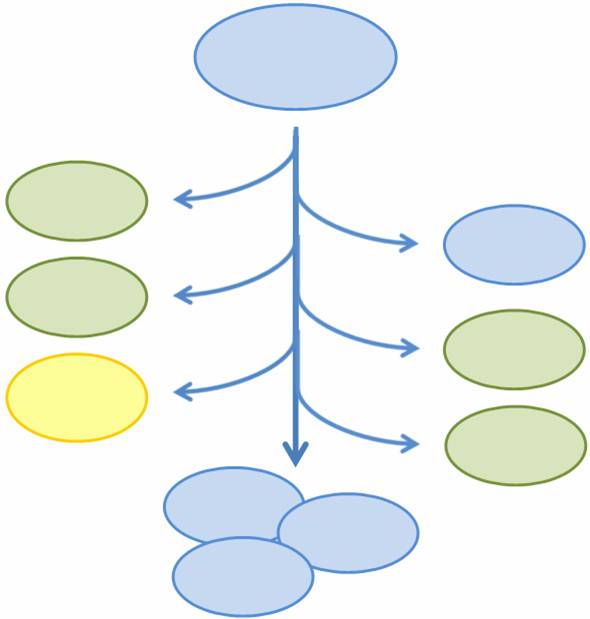
| - | In particular, we want only one of the four | + | In particular, we want only one of the four subprojects described above to be turned on in any given cell, or else the cell may be overburdened by our constructs. At the same time, we want our bacterial population to have the capability to exhibit all four functions. In addition, three of the four subprojects result in the death of the host cell through different methods of self-induced lysis. Therefore, we need a system that is able to combine all subprojects into one coherent system and that allows for self-renewal of the population. |
We propose a system in which a single bacterial strain encodes all four functions in its genome and can access each function independently through probabalistic molecular events. The engineered bacterial cells start in an undifferentiated state and randomly differentiate into one of three possible final states. To build this system, we designed two genetic devices. One relies on DNA polymerase slippage upon replication of a long stretch of short nucleotide repeats, a phenomenon termed slipped-strand mispairing (SSM). The other device uses the recombinase protein FimE to flip DNA segments. The two devices can form a system that permits the novel introduction of random multi-state differentiation into bacterial cells. Our experimental results demonstrate that short nucleotide repeats can be used as a stochastic switch and that FimE activity depends on the length of the segment being flipped. | We propose a system in which a single bacterial strain encodes all four functions in its genome and can access each function independently through probabalistic molecular events. The engineered bacterial cells start in an undifferentiated state and randomly differentiate into one of three possible final states. To build this system, we designed two genetic devices. One relies on DNA polymerase slippage upon replication of a long stretch of short nucleotide repeats, a phenomenon termed slipped-strand mispairing (SSM). The other device uses the recombinase protein FimE to flip DNA segments. The two devices can form a system that permits the novel introduction of random multi-state differentiation into bacterial cells. Our experimental results demonstrate that short nucleotide repeats can be used as a stochastic switch and that FimE activity depends on the length of the segment being flipped. | ||
{{!}}} | {{!}}} | ||
}} | }} | ||
Latest revision as of 20:49, 29 October 2008
|
People
|
Subprojects
Note: Click on the subproject title or picture for a detailed description of the subproject
Oxidative Burst
Phage Pathogen Defense
Lactose Intolerance
Vitamin Production
Population Variation
|
 "
"